Difference between revisions of "September 2010 GMOD Meeting"
m (→GMOD RPC API: The almost RESTful GMOD API) |
m (→Overview of current resources and update on DAS Meeting Cambridge 2010) |
||
| Line 978: | Line 978: | ||
== Overview of current resources and update on [[DAS]] Meeting Cambridge 2010 == | == Overview of current resources and update on [[DAS]] Meeting Cambridge 2010 == | ||
| + | |||
| + | {{ImageRight|Sept2010Jonathan.jpg|Jonathan Warren||}} | ||
Jonathan Warren, [[Media:DAS_Sept2010.pdf|PDF]], [[Media:DAS_Sept2010.pptx|PPT]] | Jonathan Warren, [[Media:DAS_Sept2010.pdf|PDF]], [[Media:DAS_Sept2010.pptx|PPT]] | ||
Revision as of 20:53, 11 October 2010
| September 2010 GMOD Meeting 13-14 September 2010 Cambridge, UK |
{{#icon: GMOD2010Europe300.png|Part of GMOD Europe 2010|200px|GMOD Europe 2010}} |
__NOTITLE__
This GMOD community meeting was held 13-14 September 2010, in Cambridge, UK, as part of GMOD Europe 2010, which also included Satellite Meetings, an InterMine Workshop, and a BioMart Workshop. The meeting was sponsored and hosted by the Cambridge Computational Biology Institute at the University of Cambridge.
GMOD Meetings are a mix of user and developer presentations, and are a great place to find out what is happening in the project, what's coming up, and what others are doing. The January 2010 GMOD Meeting was the previous event. The next meeting is likely to be held in spring 2011.
Contents
- 1 Registration
- 2 Guest Speaker
- 3 Agenda
- 4 Presentations
- 4.1 The State of GMOD
- 4.2 Help Desk Update
- 4.3 The Open Microscopy Environment: Open Informatics for Biological Imaging
- 4.4 PSICQUIC: The PSI Common QUery Interface
- 4.5 MolGenIS and XGAP
- 4.6 The GMOD Chado Natural Diversity Module
- 4.7 Cosmic GBrowse: Visualising cancer mutations in genomic context
- 4.8 GMOD Projects at the Center for Genomics and Bioinformatics
- 4.9 GMOD RPC API: The almost RESTful GMOD API
- 4.10 Overview of current resources and update on DAS Meeting Cambridge 2010
- 4.11 InterMine: new Mines and new features
- 4.12 Literature Curation in GMOD
- 4.13 Towards a GO Annotation Tool: Curation Accelerator Software
- 4.14 BioPivot: Applying Microsoft Live Labs Pivot to Problems in Bioinformatics
- 4.15 CRAWL (Chado RESTful Access Web-service Layer)
- 4.16 Lessons the GMOD community can glean from the Apache Software Foundation
- 4.17 Lightning Talks
- 5 Participants
- 6 Logistics
- 7 Sponsor: Cambridge Computational Biology Institute
- 8 Feedback
Registration
The GMOD Meeting had a registration fee (£50 early, £65 late) to cover catered lunches, coffee/tea breaks, and other expenses.
Guest Speaker
The Open Microscopy Environment: Open Informatics for Biological Imaging
- Professor, Wellcome Trust Centre for Gene Regulation and Expression, University of Dundee
- Principal Investigator, Open Microscopy Environment (OME)
The meeting's guest speaker was Prof Jason Swedlow, who discussed his work with with the Open Microscopy Environment (OME), an open international consortium that develops and releases data specifications and management tools for biological imaging. OME metadata enables image sharing, analysis, and integration with other data types.
Dr Swedlow is a Professor at the Wellcome Trust Centre for Gene Regulation and Expression and the University of Dundee. Jason's research focuses on mechanisms and regulation of chromosome segregation during mitotic cell division.
Agenda
If you are a speaker please either upload your slides, or send them to Dave Clements and he will upload them for you.
Monday, 13 September
| Time | Topic | Presenter(s) | Links |
|---|---|---|---|
| 09:15 | Introductions | Scott Cain | |
| 10:00 | The State of GMOD | Scott Cain | PDF, Summary |
| 10:30 | Break | ||
| 11:00 | Help Desk Update | Dave Clements | PDF, PPT, Summary |
| 11:30 | Keynote: The Open Microscopy Environment: Open Informatics for Biological Imaging | Jason Swedlow | PDF, PPT, Summary |
| 12:30 | Catered Lunch | ||
| 13:45 | PSICQUIC: The PSI Common QUery Interface | Bruno Aranda | PDF, Summary |
| 14:15 | MolGenIS and XGAP | Morris Swertz | PDF, Summary |
| 14:45 | The GMOD Chado Natural Diversity Module | Bob MacCallum | PDF, PPT, gdoc, Summary |
| 15:15 | Break | ||
| 15:45 | Cosmic GBrowse: Visualising cancer mutations in genomic context | David Beare | PDF, PPT, Summary |
| 16:15 | GMOD Projects at the Center for Genomics and Bioinformatics | Chris Hemmerich | PDF, PPT, Summary |
Tuesday, 14 September
| Time | Topic | Presenter(s) | Links |
|---|---|---|---|
| 09:15 | GMOD RPC API: The almost RESTful GMOD API | Josh Goodman | PDF, Summary |
| 09:45 | Overview of current resources and update on DAS Meeting Cambridge 2010 | Jonathan Warren | PDF, PPT, Summary |
| 10:15 | InterMine: new Mines and new features | Richard Smith | PDF, Summary |
| 10:40 | Break | ||
| 11:00 | Literature Curation in GMOD | Daniel Renfro | PDF, PPT, Summary |
| 11:30 | Towards a GO Annotation Tool: Curation Accelerator Software | Helen Field | PDF, KEY, Summary |
| 12:00 | BioPivot: Applying Microsoft Live Labs Pivot to Problems in Bioinformatics | Steve Taylor | PDF, PPT, Summary |
| 12:30 | Catered Lunch | ||
| 13:45 | CRAWL (Chado RESTful Access Web-service Layer) | Giles Verlarde | PDF, Summary |
| 14:15 | Lessons the GMOD community can glean from the Apache Software Foundation | Summary | |
| 14:45 | Lightning talks | Summary | |
| 15:15 | Break | ||
Wednesday & Thursday, 15-16 September
GMOD Europe 2010 continued after the GMOD meeting, starting with the Satellite Meetings and the InterMine Workshop, and finishing with the BioMart Workshop. See GMOD Europe 2010 for a complete schedule.
Presentations
This page or section is under construction.
Summaries of presentations will be posted here over the coming weeks.
The State of GMOD
GMOD is:
- A set of interoperable open-source software components for visualizing, annotating, and managing biological data.
- An active community of developers and users asking diverse questions, and facing common challenges, with their biological data.
These two things are equally important.
GMOD is used by
- hundreds of organizations
- large and small
- corporate and academic
- all over the world
- across the tree of life
What's New
- Releases
- 1.70, 2.14
- Features
- Rubberband region selection
- Drag and drop track ordering
- Collapsible tracks
- Popup balloons
- Allele/gentotype frequency
- Geolocation popups
- Circular genome support (1.71)
- Asynchronous updates (2.0)
- User authentication
- Multiple server support (2.0)
- SQLite, SAMtools (NGS) adaptors
- GMOD's 2nd Generation Genome Browser
- It's fast
- Completely new genome browser implementation:
- GBrowse based comparative genomics viewer
- Shows a reference sequence compared to 2+ others
- Can also show any GBrowse-based annotations
- Syntenic blocks do not have to be colinear
- Can also show duplications
- Chado is the GMOD schema; it is modular and extensible, allowing the addition of new data types “easily.” Covered data types in ontologies, organisms, sequence features, genotypes, phenotypes, libraries, stocks, microarrays, with natural diversity recently being rolled into the schema (but not yet released).
- 1.0 Release solidified the Chado that most people were already using from source.
- 1.1 Introduced support for GBrowse to use full text searching and “summary statistics” (ie, feature
density plots). Version 0.30 of Bio::DB::Das::Chado is needed for these functions.
- New (2009) web front end for Chado databases
- Set of Drupal modules
- Modules approximately correspond to Chado modules
- Easy to create new modules
- Includes user authentication, job management, curation support
- A MediaWiki extension (MediaWiki software used at Wikipedia, GMOD.org)
- Provides graphical user interface (GUI) to wiki tables
- Can also provide GUI to database tables
- Work in progress to use this with Chado
- Potential to give wiki access to a Chado database
- See http://ecoliwiki.net
- BioMart is a query-oriented data management system
- Provides a web based query interface
- Strong data federation
- BioMart Workshop on Thursday.
- InterMine is a query-oriented data management system
- Provides a web based query interface
- Very flexible queries and query optimization
- InterMine Workshop on Wednesday
- Genome annotation pipeline for creating gene models
- Output can be loaded into GBrowse, Apollo, Chado, …
- Incorporates
- SNAP, RepeatMasker, exonerate, BLAST, Augustus, FGENESH, GeneMark, MPI
- Other capabilities
- Map existing annotation onto new assemblies
- Merge multiple legacy annotation sets into a consensus set
- Update existing annotations with new evidence
- Integrate raw InterProScan results
- Maker Online in beta
- Java-based GUI application for browsing and annotating genomic sequences
- Can be installed via WebStart (ie, by clicking on a link)
- Can read/write to Chado, GFF3, GenBank, GAME XML
Next GMOD Meeting?
- Next Spring Sometime:
- ABRF: Association of Biomolecular Resource Facilities
- Feb. 19-22, San Antonio, TX
- Biology of Genomes
- May 10-14, Cold Spring Harbor Lab, NY
- Suggestions?
Help Desk Update
Mailing List Archives
GMOD Mailing Lists are all over. Many are hosted at SourceForge, but several are elsewhere (EBI, Bluehost, Berkeley, ...). Some don't have public archives and those that do are spread around The lists at SourceForge have searchable archives but the search interface is frustrating.
Since May/June 2010, all emails to GMOD mailing lists have been archived in a single searchable hierarchy at Nabble. Nabble has a functional search capability and you can now search all lists, or just a single list.
GMOD Membership Requirements
GMOD's requirements for software to join GMOD were codified in February 2010, following January 2010 GMOD Meeting]]. These requirements were in use before February, but were inconsistently applied.
Version 1 Requirements:
- Meets a common need
- Useful over time
- Configurable and Extensible
- Open source license for all users
- Interoperable with existing GMOD components
- Commitment of support
For next version, want to add:
- Support mailing list that is publicly archived
- Publicly accessible code repository
Discussion favored these additions. The issue of incompatible open source licenses also came up. GMOD currently requires any OSI approved license. However, some of those licenses are not compatible with each other, meaning they such components can't be bundled together.
GMOD Promotion
Help spread the word about GMOD components and the GMOD project.
- Why?
- Increased visibility leads to
- → Increased adoption, which leads to
- → more projects contributing back
- → Increased adoption, which leads to
- Increased adoption & development leads to
- → increased funding
- How?
- Cite GMOD, GMOD Components in your papers, presentations, grants
- Powered by GMOD icons
- Speakers at your event; not just Scott and Dave. PIs and developers are also available.
- Graphics & slides for your presentations, posters
- Presentation and event promotion
- Brochures (GMOD project, events)
- Bling!
The GMOD Promotion page launched in July 2010.
GMOD Logo Program
Nine projects got new logos in the Spring 2010 Logo Program. Logos were done by John Aikman's Spring 2010 Advanced Design class at Linn-Benton Community College, Albany, Oregon, United States. Each project worked with 2-3 students during the quarter to produce the selected logos.
We might do this again in 2011.
2010 GMOD Community Survey
The 2008 GMOD Community Survey covered components and project wide topics. The 2009 GMOD Community Survey focused on genome and comparative genomics browsing. The 2010 GMOD Community Survey will cover components and project wide topics. We may use it to produce a GMOD Project publication.
These surveys help guide the project and also show potential and current GMOD users what the larger community is doing.
Look for the 2010 survey in October.
Events
The satellites at the January 2010 GMOD Meeting were such a success that we decided to do them again. Satellites are birds of a feather discussions where participants with a common interest discuss that topic. The satellites at this meeting were:
See the satellite meeting pages for summaries of the discussion.
In 2010 we held our 4th summer school in May at NESCent, in Durham, North Carolina, US. We had 62 applicants for 25 slots.
The 2011 course will likely be at NESCent again. However, starting in 2011, summer school expenses will no longer be covered by a grant (see below). This means that we will start charging tuition, and that we will also start seeking sponsors.
Summer school sessions become online tutorials that include starting and ending VMware images, step by step instructions, and example datasets.
- Other Upcoming Events of Note
- Biocuration 2010
- October, Tokyo, Japan
- Pathway Tools Workshop
- October, Menlo Park, California, US
- GMOD Evo Hackathon
- November, Durham, North Carolina, US
- Computational and Comparative Genomics
- November, Cold Spring Harbor, New York, US
- Plant and Animal Genome
- January, San Diego, California, US
- Workshop on Molecular Evolution
- January, Cesky Krumlov, Czech Republic
- Galaxy Developers Conference
- 2011, Europe
JBrowse Development
- 1.1 just released
- Scalability: very large data sets, including NGS reads, human EST/SNP tracks
- Extensibility: custom tracks
- Backward incompatible JSON format
- 1.2 Release (December 2010)
- improved NGS display (paired-end reads, possibly read-to-genome alignments)
- reduced memory usage for NGS
- minor UI enhancements including y-axis labels for wiggle tracks
JBrowse Grant Proposal
Sent proposal in this summer; if approved will start around February 2011.
- JBrowse concepts have proven themselves
- Scalable to coming data set sizes
- GBrowse development will wind down during the grant.
- New Features
- JBrowse ecosystem on par with what GBrowse has
- DAS and web services support
- Scalability and NGS
- Large numbers of tracks
- Community annotation (upload/publish, tagging, comment, …)
- Mobile device support?
- GBrowse → JBrowse Migration Support
- Migration Scripts: Config files, data (data is easy)
- Simultaneous GBrowse and JBrowse support
- JBrowse running on top of GBrowse config and data
New Components
- ISGA
- Chris Hemmerich et al. at Indiana U.
- Bioinformatics pipeline service software built on Ergatis
- Newest GMOD component
- WebGBrowse
- Ram Podicheti et al. at Indiana U.
- SOBA
- Ginger Fan et al. U of Utah
- GFF3 file analysis and reporting
- Tabular and graphical reports
- Nominated and approved, code being refactored
- GMOD-DBSF, genes4all, …
- Alexie Papanicolaou at CSIRO
- Drupal based toolkit for building organism web sites
- Submitted for publication; not yet nominated
Some Interesting Documents
- How to load a Chado Database into BioMart
- AO Keliet, J Amselem, S Derozie, and D Steinbach, all @ INRA URGI
- Choosing a genome browser for a Model Organism Database
- surveying the Maize community
- TZ Sen, LC Harper, ML Schaeffer, CM. Andorf, TE Seigfried, DA Campbell, and CJ. Lawrence
- How and why MaizeGDB picked GBrowse
- Appeared in Database: The Journal of Biological Databases and Curation
- Nature Methods Supplement on visualizing biological data, March 2010
- Visualizing biological data - now and in the future
- SI O'Donoghue, et al.
- Visualizing genomes: techniques and challenges
- CB Nielsen, et al.
- Visualization of multiple alignments, phylogenies and gene family evolution
- JB Proctor, et al.
- Visualization of image data from cells to organisms
- T Walker, et al.
- Visualization of macromolecular structures
- SI O'Donoghue, et al.
- Visualization of omics data for systems biology
- N Gehlenborg, et al.
GMOD on the Web
- GMOD.org
- Moving from CSHL to OICR, real soon now
- MediaWiki upgrade
- Probably lots of new extensions
- Maybe a modified skin
- Look into adding
- User log section
- Scrapbook for contributed code
- Membership directory (TableEdit based)
- Semi-automated publication listing/linking
- Should GMOD have a social presence?
GMOD already has mailing lists, wiki, GMOD News (RSS), and IRC. Should GMOD have a presence in social media as well? If so, what should the goals be? Outreach? Community building or forums? Social bookmarking? Which tools should we use: Twitter, Facebook, Connotea, StumbleUpon, Technorati, Nature Network…
ISB uses Connotea to bookmark "biocuration", "text mining", and "semantic annotation" papers.
This generated some discussions and some conclusions:
- Community bookmarking may be worthwhile.
- If you can automatically tweet page updates and news items, do it.
- Don't manually post stuff to twitter
- Don't build community through Facebook. There are better time investments.
The Open Microscopy Environment: Open Informatics for Biological Imaging
PSICQUIC: The PSI Common QUery Interface
The Proteomics Standards Initiative (PSI) Common Query Interface (PSICQUIC, pronounced like "psychic" - most of the time) standardizes access to molecular interaction data. PSICQUIC is a web service specification based on PSI standards. Resources that implement PSICQUIC are listed in a public registry. There are currently more than 14 million binary interactions from at least 12 different resources (IntAct, Reactome, chEMBL, ...) available using PSICQUIC. This widespread adoption allows client programs that speak PSICQUIC to uniformly access all this no matter where it is located.
PSI talked for many years about standards and formats and how to share data. They 2002-2006 thinking about standards. They found it was very complicated to agree on something. but that it has been easy to implement. Most PSICQUIC implementation came out of 3 biohackathons.
PSICQUIC Web Services
- Methods
Several methods are supported:
- getByInteraction - Retrieves interactions by using an interaction AC.
- getByInteractionList - Retrieves interactions by using a list of interaction AC.
- getByInteractor - Retrieves interactions by using a participant identifier.
- getByInteractorList - Retrieves interactions by using a list of participant identifiers.
- getByQuery - Retrieves interactions by using a Molecular Interaction Query Language (MIQL) query (full text searches)
- getVersion - Returns the version of the web service implementation.
- getSupportedDbAcs - Returns the supported database identifiers
- getSupportedReturnTypes - Returns the list of available format types for the results.
A limited number of interactions can be fetched. It is possible to retrieve large datasets using pagination. Most methods have two additional parameters:
- First result: Index for the first result to retrieve.
- Max results: Number of interactions returned per query.
IMX Consortium and UniProt identifiers are currently being used. Don't have the one single identifier yet.
- SOAP and REST
As PSICQUIC is a Web Service, you can access the data:
- Via SOAP
- A WSDL file exists, and it is the same for all the databases.
- IntAct has developed a Java client, but any other language can be used.
- The SoapUI client uses this.
- However, SOAP's future in PSICQUIC is uncertain and may go away in the future.
- Via REST
- Retrieving data directly by using a URL
- Easy to access and data can be obtained just using an internet browser.
- Effective for scripting.
Formats
PSICQUIC has two standard formats: PSI-MI XML and PSI-MI TAB. The XML is more complete, and therefore more verbose. PSI-MI TAB is a tabular format.
- Try these queries at IntAct:
Other formats are in progress:
- BioPAX (IntAct example)
- rdf-xml (IntAct example)
- rdf-n3 (IntAct example)
- rdf-n3-triple (IntAct example)
- rdf-turtle (IntAct example)
As these formats are works in progress, some of these links may fail.
- PSICQUIC Registry
The PSICQUIC registry contains a list of the PSICQUIC services available from different providers. It is a web service itself, and it can be accessed remotely using REST. Information can be found about the services, such as the URLs to use, number of interactions provided, versioning, etc. The registry classifies the different services with tags from a PSI ontology. Querying by tags is a work in progress. Instructions on using the registry are at Google Code.
MIQL
PSICQUIC also defines the Molecular Interactions Query Language (MIQL). MIQL allows more powerful and flexible queries and is the default query syntax for PSIQCUIC. Designed for fast and effective searches on PSI-MI TAB files. All fields (columns) can be searched with specific queries. MIQL is a consensus between the different databases, so you should be able to use the same query across different repositories.
The MIQL syntax is based on the Lucene syntax. A query is broken into terms and operators:
- Terms: single words or phrases (group of words surrounded by quotes). E.g. brca2 AND “pull down”
- Fields: search in specific columns. E.g. brca2 AND species:human
- Term modifiers: wildcard searches, fuzzy searches, proximity and range searches. E.g. brc*
- Operands: OR (or space), AND, NOT, +, -. E.g.
- brca2 AND rpa1 / brca2 NOT mouse / +brca2 –mouse –expansion:spoke
- Grouping and field grouping: brca2 AND (mouse "in vitro")
Creating a PSICQUIC Service
Simplest recipe to implement PSICQUIC
- Ingredients:
- PSI-MITAB compliant file.
- Subversion: to get the source code.
- Maven: to run the scripts and start the service.
- Steps:
- Generate the MITAB compliant file.
- Get the Reference Implementation (RI)
- Run the script to index the file.
- Start the service with the script provided .
PSICQUIC Applications
PSICQUIC is already implemented in several existing applications, including Cytoscape 2.7.x, PSICQUIC View, Envision2, and PSICQUIC Client for Android.
There is not currently anything in the GMOD suite that uses PSICQUIC. Should there be?
PSICQUIC Development
- Smart PSICQUICs: Identification and removal of redundancy
- Merger and Cluster PSICQUIC services
- PSICQUIC 2.0
- Overcome the current limitations and many fancy features:
- Queries using CV terms not possible in the reference implementation (it is possible in IntAct).
- PSI-MI XML is created from the MITAB, so no n-ary interactions.
- New features:
- Redundancy detection mechanism. ROG/RIG ids by default.
- Built from PSI-MI XML, so complex data available.
- Overcome the current limitations and many fancy features:
- A GMOD component?
Flybase is using Chado Interaction format. Ecoli has lots of interaction. Can we have a Chado service that talks PSICQUIC?
Following the talk a couple of possible actions arised:
- Exporting from Chado to MITAB, so we can just create PSICQUIC services from any Chado-based application.
- Creating a component / adding interaction information to existing components.
Bruno is unfamiliar with Chado, but if someone wants to give it a shot, he is more than willing to help and participate. All information about PSICQUIC can be found at Google Code.
And some basic information about the MITAB format may help.
MolGenIS and XGAP

Morris Swertz, PDF
MolGenIS is a flexible bioinformatics application toolkit for data management and interfacing. XGAP is an •eXtensible Genotype And Phenotype system that was generated with MolGenIS to store and visualize xQTL and GWAS data.
One aim of this talk is to explore possible links between MolGenIS and GMOD: [[Chado], DAS, BioMart, InterMine, GBrowse, ...?
MolGenIS
MolGenIS has been used to generate systems for many different types of applications and datatypes. MolGenIS based systems and users include GEN2PHEN, XGAP, UMCG, FIMM, Sysgenet, and many others.
MolGenIS is a system generator. It addresses the recurring issue of generating custom databases for each new application that comes along. The traditional approach requires database design, backend (server) coding, API development, and user interface coding, all of which is bioinformatician intensive. This approach does not have reusability and interoperability as a natural byproduct of development. With MolGenIS system developers provide a system definition which MolGenIS then uses to automatically instantiate a system that implements the definition. Writing a system definition requires learning new skills, but is still much less time intensive then creating a system from scratch.
MolGenIS includes built in support for many features:
- database generation
- server code generation
- User interface generation, including edit interfaces and audit trails
- Import/Export to Excel
- R interoperability
- workflow ready web services using REST, SOAP and RDF
- UML documentation of underlying models
MolGenIS also comes with extensive documentation, including a development manual.
Generated systems can also be customized. The user interface can be extended with plugins implemented in as a Java class, and a layout definition. Similarly, plugins can be added to the server side by defining a Java class.
The database backend currently uses a custom object-relational mapping (ORM). Hibernate was considered six years ago, but was lacking key features. The long term hope is to migrate to a standard ORM such as Hibernate.
XGAP
XGAP (eXtensible Genotype And Phenotype) was developed for xQTL and GWAS data.
The data is logically in a series of matrices with a different matrix for each datatype (e.g., genotype, microarray, LC/MS, ...). The initial idea was to create a database table for each datatype, but this would have led to a proliferation of structurally similar database tables, and would require schema changes with the addition of each new type in the future. (Imagine Chado's feature table split into gene, ssr, snp, exon, etc. tables.)
XGAP addresses this by embracing a generic matrix model: any trait X any subject. All matrices are stored in a common database table where each row corresponds to a single element in a matrix. Schema changes are not required to add new matrices or new columns to existing matrices. This is all done by adding matrix and column definitions to definition tables in the database.
FuGE (Functional Genomics Experiment) is a standard model for this type of information. XGAP builds on top of this.
GMOD Link Ideas
- Chado
- XGAP harmonization towards Chado?
- MolGenIS 4 Chado? Did BioSQL a few years ago.
- BioMart / InterMine
- Consume BioMARTdata to auto-annotate experimental data?
- Export XGAP experiments into MART/MINE query environments?
OntoCAT
The GMOD Chado Natural Diversity Module

Motivation
- Manage phenotypic and genotypic data for both field collected and captive bred organisms
- Store collection site information for growing "next gen"-based variation data
- Leverage existing/future Chado modules, GMOD tools and know-how
Developmental History
- 2007
- Early version:
- HeliconiusDB @ NESCent (National Evolutionary Synthesis Center)
- Inspired by GDPDM (The Genomic Diversity and Phenotype Data Model)
- 2009-2010
- Reincarnation spearheaded by:
- Sook Jung @ Washington State University, GDR (Genome Database for Rosaceae)
- GMOD working group formed
- Reincarnation spearheaded by:
- August 2010
- Natural Diversity module merged into Chado svn trunk
Schema
Makes use to the pre-existing stock module. Adds support for Experiment, Geolocation, and Genotype and Phenotype (reusing some existing tables), The talk walked through how three specific use cases would be implemented:
- Cross experiment
- Field collection
- Phenotype assay
CV Terms and APIs
Schema is very flexible. nd_experiment.type and nd_experiment_stock.type are key. There are several ways to do the same thing. The working group is hoping to agree on core CV terms to aid API development. VectorBase is planning a simplified API that abstract the module's tables into:
- stocks
- experiments, for which we propose at least three subclasses:
- field collections
- phenotyping experiments
- genotyping experiments
- projects
- protocols
Cosmic GBrowse: Visualising cancer mutations in genomic context

The Cancer Genome Project (CGP) started in 2000. COSMIC, the Catalogue Of Somatic Mutations In Cancer was launched on 4 February 2004. COSMIC is a website and backing Oracle database. COSMIC mutation data comes from several sources.
- Three curators who read and annotate publications.
- Other database(s) e.g. TP53 (IARC), International Agency for Research on Cancer
- Sequencing/mutation detection
The project is planning on launching COSMIC GBrowse on 22 September 2010.
- GBrowse and CGP
| Q. | How could we visualise the data deluge from next generation sequencing? |
|---|---|
| A. | GBrowse. (See [Keiran Raine's presentation at the January 2010 GMOD Meeting.) A near instant solution to the problem (days/weeks, rather than months/years for an in house solution). Looked at lots of options. GBrowse looked like the clear winner - it's configurable and meets needs. |
| Q. | COSMIC was designed to be gene centric but what about sequencing whole cancer genomes and visualising mutations in genomic context? |
| A. | Gbrowse. Again! |
Data
- Reference
- Reference genome (GRCh37) + cytogenetic bands
- Ensembl annotations (e! 58)
- Cosmic Transcripts
- Cosmic
- Mutations (substitutions, insertions/deletions)
- Rearrangements
- Copy Number Profiles
- analysis of SNP6 microarray data over 800 cell lines
- % samples which have copy number features (amplification, homozygous deletion, LOH, change)
Configuration and Setup
- Hardware
- 5 Virtual Machines [Debian Linux, 2G RAM) ]
- dev + master + renderfarm slaves (2) + PostgreSQL. The Master talks to the two slaves, both of which talk to the reference and mutations databases.
- Software
- apache 2.2.9
- mod_fastcgi 2.4.6
- GBrowse 2.13 (perl 5.10.0 + BioPerl 1.61 + Bio::Graphics 2.11]
- Note:' 'There was significant renderfarm development between 2.13 and 2.14
- Databases
- PostgreSQL
- 2 databases: ‘Reference’ and ‘Cosmic’
- scripts to query/format/populate these databases
- PostgreSQL
- Configuration
- cosmic css/theme
- perl callbacks: glyphs, colours, hyperlinks, popups/tooltips
Display
COSMIC GBrowse shows:
- genes, COSMIC transcripts, non-coding RNA
- breakpoints with lightning (!) and detailed popups
- Copy number change, with color, and links to CONAN.
- LOH, with color
- Mutations density
- Mutation details (intronic, nonsense, missense, Silent, Non-coding, frameshift, in frame, complex, deletion, insertion), with colors and shapes, provide a key and detailed popups
- See slides for screenshots.
Future Development
- At COSMIC
- Embed cosmic GBrowse in some cosmic web pages - replace old and slow drawing code and extend functionality.
- Current version is a summarised view of whole cosmic dataset. We need to be able to display subsets of data. How can we display all mutations for a specific sample or group of samples, or from a specific tissue or tumour type? oo many for a static list of data sources, but there is a neat trick ..
- Define data source in the URL, eg sample COLO-829: http://www.sanger.ac.uk/fgb2/gbrowse/sample_COLO-829
- GBrowse.conf ... (need at least 2.09)
[=~sample_.+] description = Cosmic Database v48 (sample filtered) path = /gbrowse/bin/source_config.pl -sample $1 | # path points to a script which generates the config # sample name ‘COLO-829’ is passed to the script from regular expression # track configuration generated for data source COLO-829 … [Mutations] remote feature = http://…/cosmic_export.cgi?sample=COLO-829 # cgi script returns COLO-829 mutation data from COSMIC
- GBrowse Developement
- remote feature - perl callbacks cannot be used until Safe::World is fixed
- init_code - perl callbacks defined with init_code not accessible from slaves
- BAM/SAM read sorting by similarity to reference
- GC plots can give >100% values
- CGP
CGP committed to using GBrowse as its internal browser for next gen sequencing data, and an external browser for COSMIC data (genomic view of mutations, breakpoints and copy number data). COSMIC GBrowse to be released soon (22/9/2010?). CGP is also involved in GBrowse development. A new developer has been recruited, but details are still being discussed.
GMOD Projects at the Center for Genomics and Bioinformatics

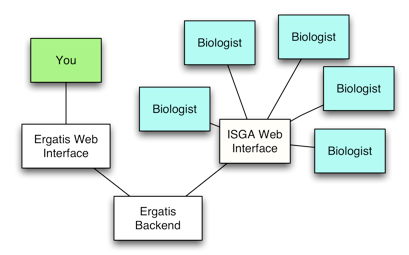
A Simple Web Interface for Configuring GBrowse: WebGBrowse
(By Ram Podicheti, as channeled by Chris)
WebGBrowse is a web interface for configuring GBrowse installations. You can upload GFF files and optionally upload an existing GBrowse config file to use as starting point. From there, you can add, edit, and remove new tracks using web forms. WebGBrowse comes with extensive help embedded in the forms and includes a tutorial. Users can preview their changes at any point in GBrowse. WebGBrowse makes GBrowse more feasible for small projects who can figure out configuration, but don't have the resources to setup their own server.
WebGBrowse can be downloaded and locally installed. There is a mailing list for support, feature requests, and contributions. We want to help you help us add support for more features. WebGBrowse has passed the nomination process and is now a pending GMOD component. Waiting only migration of development environment to a public repository.
WebGBrowse has support GBrowse 2 for quite a while. It does not support callbacks yet (and this is hard due to security considerations),
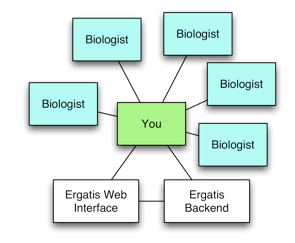
Web-based Bioinformatics Pipelines for Biologists: ISGA
(By Chris, Aaron Buechlein, Ram, Jeong-Hyeon Choi, and Boshu Liu as channeled by Chris)
ISGA is a workflow management system that can meet the needs of a small sequencing center. It supports flexible pipeline definition for new pipelines, and for incorporating new programs as components. ISGA supports distributed computing environments, if you have a potential need to grow beyond local computing resources. ISGA was created at CGB to minimize CGB staff involvement in running pipelines. ISGA frees up staff resources for building new pipelines.
ISGA is built on top of Ergatis. Ergatis is developed and support by the Institute for Genome Sciences, U. Maryland. Ergatis enables building pipelines from existing programs, supports distributed computing environments, and has robust monitoring of pipeline execution. Ergatis comes with 10+ readily available pipelines, and there are more available in the community. There are currently 220 tool/component definitions that come with Ergatis, and again, there are more in the community. Components and pipelines are defined in XML. XML/BSML is the common data exchange format. XML/BSML is optional, but recommended for reusable components. Includes conversion tools for FASTA, GFF, Chado, etc... This isolates format changes from other programs. Ergatis runs on Condor out of the box.
Ergatis's interface assumes that a computationally savvy biologist will be using it. In practice, this can lead to the informatics staff being the practical interface between biologists and Ergatis. CGB had several goals when developing ISGA:
- Wanted to support single-lab biologists that are self-sufficient but have limited bioinformatics resources and that embrace tools that don’t require extensive training
- Ability for biologists to run pre-configured pipelines quickly
- Option to customize specific tools in a pipeline
- An interface that encourages exploration:
- Remove complexity and information biologists don’t need
- Inline help
- Immediately detect errors and allow biologists to correct them
- Return output in useful formats
- Simple tools for visualizing and searching large result sets
ISGA does this and several other things too: First, it simplifies pipelines by hiding housekeeping components and by grouping components into clusters representing processes. ISGA supports customization. Users can disable components, replace components with pre-computed data, and edit scientifically-active program parameters. It also provides help and validation for all forms, and incorporates visualization and analysis tools. In addition ISGA support the concepts of users and data privacy, and users can upload and download data,
Why develop ISGA as a separate package?
ISGA only re-implements the web interface of Ergatis. Ergatis libraries, component definitions, and method of running and monitoring pipelines is used by ISGA as-is. ISGA adds and removes Ergatis features such as accessing component information and building pipelines from components. ISGA biologist users need to be given limited functionality for simplicity and security. Ergatis bioinformatician users need full functionality and a complex interface to work efficiently. A hybrid ISGA/Ergatis interface wouldn’t serve anyone.
Present and Future
ISGA at Indiana has run over 100 pipelines, and has more than 60 users. There are two external sites evaluating their own ISGA installation that CBG knows of.
Recent developments in ISGA include
- Celera assembly pipeline
- Ability to accept parameters with pipeline inputs
- Ability to iterate components over a list of pipeline inputs
- Conversion scripts for Hawkeye visualization
- Installation instructions :shame
- isga-users@lists.sourceforge.net
- Administration improvements
- Online configuration
- User classes and pipeline quotas
And there is more in the works:
- Pipelines
- SHORE SNP Calling (ISGA)
- Gene clustering over Microbial phylogenies (Ergatis)
- Transcriptome annotation pipeline (Ergatis)
- Methyl-seq (Ergatis)
- Features
- Pipeline reproducibility and provenance
- User groups and sharing
- Modular pipeline and toolbox installation
- ISGA pipelines as standalone Ergatis templates
- ISGA pipeline over Amazon EC2 via CLoVR
- CloVR
- Cloud Resources through CloVR
- Execute Ergatis Pipelines over an SGE instance hosted on Amazon EC2 machine images
- CloVR manages creation and shutdown of cloud images as part of pipeline
- Upload input as part of pipeline or access data hosted at Amazon
- Results are retrieved to local machine
- Ergatis assumes a shared filesystem, so some modification is required to manage file transfers
- Using CloVR with ISGA
- ISGA/Ergatis pipelines can be ported to ISGA/CloVR
- ISGA installation communicates with local Ergatis and CloVR
- EC2 presents challenges for billing customers
GMOD RPC API: The almost RESTful GMOD API

Josh started with this scenario:
Fetch me all genes annotated with GO:0003677 (DNA Binding) from D. melanogaster, C. elegans, T. castaneum, and B. mori. Then fetch the current ID, symbol and list of orthologs for each.
We currently do this with a mixture of file downloads, SQL calls to different DB systems, a patchwork of parsing scripts, and screen scraping. Instead, we should be doing:
$ curl http://flybase.org/gmodrpc/v1.1/ontology/gene/GO:0003677 $ curl http://wormbase.org/gmodrpc/v1.1/ontology/gene/GO:0003677
This idea was motivated by a discussion at the July 2008 GMOD Meeting where a simple request, like the one above, required screen scraping. This work uses the REST protocol to gather information. REST is an alternative or successor to CORBA, a heavyweight protocol for sharing information, and SOAP, a more recent, but still too heavy for our purposes protocol for doing the same.
The GMOD RPC API proposal supports a number of information services:
- Organisms
- Full text search
- Location
- Gene ontology
- Orthology
- Gene
- Organism
- Fetch common gene page
In an ideal world each MOD would provide these services.
The idea is to provide top level classes. FlyBase will provide a specific Chado/Perl based implementation. However, the proposal is trying to be agnostic in terms of what data types are expected. Josh is working on the Perl implementation. Others are working on PHP (Jim Hu) and Java implementations.
Perl Implementation
- Strict MVC separation.
- Moose used for the model
- Moose is much better than Perl 5 objects.
- GMOD RPC API will provide base code and utility functions. You extend base class of each service to implement based on your environment.
- Template::Toolkit for the view
- Perl’s Dancer for the controller
- Simple and clean with minimal dependencies. Perl implementation of Ruby's Sinatra.
- Easy to install. Decided against Catalyst because of installation and dependencies. Want something simple to get this off the ground.
- Can be run under CGI, PSGI (Plack), and FastCGI on a variety of web servers (Apache, Nginx and lighttpd)
- Moose used for the model
- Log::Log4perl for logging
- Standard Test::More unit tests
Goals
- Short term
- Alpha release by end of October 2010
- Beta release by end of December 2010
- Long term
- DAS tie in
- Validation for XML formats
- Java, PHP and Python APIs
- Evaluate additional API features
How to participate
- Subscribe to gmod‐devel
- GMOD REST API
- SVN code repos
Discussion
Plan is to keep old version APIs around. That is to keep the old URLs accessible by having constant and stable URLs.
There is a mechanism to query what services are available - returned as an XML list of services from above list (organism, ...).
Queries can use taxonomy id, or just genus and species. Can get sequence by asking for a gene and SO terms
Can this REST interface be made a standard feature in a GBrowse implementation? That's ideally what we should shoot for. This is where this should go.
Is there a way to pull this in whole, instead of at a retail level? Not currently, but FlyBase puts gene reports into XML. Then use XSLT to generate the web pages, and then save XML in Lucene for ful text searching.
Overview of current resources and update on DAS Meeting Cambridge 2010

InterMine: new Mines and new features
Literature Curation in GMOD
Towards a GO Annotation Tool: Curation Accelerator Software
BioPivot: Applying Microsoft Live Labs Pivot to Problems in Bioinformatics
CRAWL (Chado RESTful Access Web-service Layer)
A programmatic interface for querying pathogen genomics data
Lessons the GMOD community can glean from the Apache Software Foundation
Lightning Talks
Participants
| Participant | Affilliation(s) | URL |
|---|---|---|
| Scott Cain | OICR | http://gmod.org/ |
| Dave Clements | NESCent, GMOD | http://nescent.org http://gmod.org |
| Josh Goodman | FlyBase - Indiana University | http://flybase.org |
| Richard Smith | Cambridge University | http://www.intermine.org |
| Anup Mahurkar | Institute for Genome Sciences University of Maryland School of Medicine | |
| joan pontius | SAIC-NCI-FREDERICK Laboratory of Genomic Diversity | http://lgd.abcc.ncifcrf.gov/cgi-bin/gbrowse/cat/ |
| Christelle Robert | The Roslin Institute The University of Edinburgh | |
| Matthew Eldridge | Cancer Research UK - Cambridge Research Institute | |
| Fengyuan Hu | Department of Genetics, University of Cambridge | |
| Daniel Renfro | EcoliWiki, SubtilisWiki, Hu lab - Texas A&M University | EcoliWiki, SubtilisWiki, GONUTS |
| Ellen Adlem | Cambridge University Cambridge Institue of Medical Research | http://www.t1dbase.org |
| Kerstin Koch | KWS Saat AG Bioinformatics Grimsehlstr. | |
| Oliver Burren | Cambridge University | http://www.t1dbase.org |
| Chris Jiggins | University of Cambridge | http://heliconius.zoo.cam.ac.uk/ |
| Jason Swedlow | Wellcome Trust Centre for Gene Regulation and Expression, University of Dundee, The Open Microscopy Environment (OME) | http://gre.lifesci.dundee.ac.uk/staff/jason_swedlow.html, http://www.openmicroscopy.org/ |
| Dave Beare | Cancer Genome Project, Wellcome Trust Sanger Institute | http://www.sanger.ac.uk/research/projects/cancergenome.html |
| seth redmond | Imperial College / Vectorbase | |
| Chris Hemmerich | http://cgb.indiana.edu | |
| Emmanuel Quevillon | Institut Pasteur | http://www.pasteur.fr/ip/easysite/go/03b-00000m-0q8/recherche/logiciels-et-banques-de-donnees |
| Bob MacCallum | VectorBase Imperial College London | http://www.vectorbase.org |
| Ewan Mollison | Tun Abdul Razak Research Centre, Hertford | http://www.tarrc.co.uk |
| Jen Harrow | Wellcome Trust Sanger Institute | |
| Gos Micklem | University of Cambridge | http://www.sysbiol.cam.ac.uk/index.php?page=dr-gos-micklem |
| Malcolm Hinsley | Wellcome Trust Sanger Institute | |
| Gemma Barson | Wellcome Trust Sanger Institute | http://www.sanger.ac.uk/ |
| Brett Whitty | Michigan State University | http://buell-lab.plantbiology.msu.edu, http://solanaceae.plantbiology.msu.edu, http://potatogenome.net |
| Morris Swertz | Genomics Coordination Center, University Medical Center Groningen EMBL - European Bioinformatics Institute | http://www.molgenis.org |
| Jerven Bolleman | UniProt Swiss-Prot | |
| Alex Kalderimis | InterMine, Cambridge University | http://www.intermine.org, http://www.flymine.org |
| Oksana Riba Grognuz | Swiss Institute of Bioinformatics (SIB) Department of Ecology and Evolution, University of Lausanne | |
| Dr Helen Imogen Field | FlyBase Dept Genetics University of Cambridge | http://www.gen.cam.ac.uk/research/flybase.html |
| Kim Rutherford | Cambridge Systems Biology Centre | http://www.pombase.org/ |
| Robert Wilson | National Institute for Medical Research, London | |
| Gerd Anders | Public research institute: Max-Delbrueck-Centrum Berlin (MDC), Researcher and database developer | http://www.mdc-berlin.de/en/research/core_facilities/cf_massspectromety_bimsb/teammember/index.html http://www.mdc-berlin.de/en/research/core_facilities/cf_bioinformatic/teammember/index.html |
| Joeri van der Velde | University of Groningen, GBIC UMGC, dept. of Genetics Genomics Coordination Center | |
| Jonathan Warren | The Sanger Institute | http://www.dasregistry.org |
| Stephen Taylor | CBRG, Oxford University | http://www.cbrg.ox.ac.uk/ |
| Bruno Aranda | EMBL-EBI | http://www.ebi.ac.uk/intact, http://psicquic.googlecode.com |
| Mahmut Uludag | European Bioinformatics Institute | |
| Giles Velarde | The Sanger Centre | http://www.genedb.org, http://www.sanger.ac.uk |
| Andy Jenkinson | European Bioinformatics Institute | |
| Kevin Howe | Wellcome Trust Sanger Institute |
Logistics
This meeting was held in the Biffen Lecture Theatre, in the Department of Genetics on the University of Cambridge campus.
Wireless
Thanks to Ian Clark, the Biffen Lecture Theatre had wireless. From the Cambridge website:
Members of the University of Cambridge can either use their Raven login to connect to Lapwing or they can configure their computer to use Eduroam. Visitors from institutions participating in the Eduroam initiative can also use Eduroam, but should obtain instructions from their home institution.
Visitors who cannot use Eduroam for any reason can obtain a time-limited Lapwing ticket by asking their contact in Genetics to mail the following information to the CO:
Accounts were setup for all attendees for the duration of GMOD Europe 2010.
Power
The Biffen Lecture Theatre has wireless, but it does not have power outlets throughout the room.
To help us through the days, Gos Micklem secured a 15-socket extension strip which was placed at the back of the room. Please come to the meeting fully charged.
Transportation and Lodging
See the Transportation and Lodging sections on the GMOD Europe 2010 pages for details.
Sponsor: Cambridge Computational Biology Institute
The September 2010 GMOD Meeting was sponsored by the Cambridge Computational Biology Institute, which is hosting the meeting and is also the home of InterMine. The CCBI is "set up to bring together the unique strengths of Cambridge in medicine, biology, mathematics and the physical sciences. Its aim is to create a centre of excellence in research and teaching and to promote collaborations both within the Cambridge area and beyond."
Please thank Gos Miclem, Shelley Lawson, and Richard Smith for hosting the event. We could not have done this without their support, effort and time.
Feedback
Please provide your feedback! We will use it to guide future GMOD events.