Difference between revisions of "BioMart Tutorial 2011"
Anthony Cros (Talk | contribs) |
(→4. Configuring mart (MartConfigurator)) |
||
| Line 221: | Line 221: | ||
Double-clicking on the access point icon will open a new window that allows you to modify the access point. | Double-clicking on the access point icon will open a new window that allows you to modify the access point. | ||
References | References | ||
| + | |||
| + | = 5. Querying a BioMart server via REST API = | ||
| + | |||
| + | == 5.1 MetaData queries == | ||
| + | |||
| + | == 5.2 Data query == | ||
| + | |||
| + | |||
To read more about BioMart, refer to the recent articles describing the BioMart software and its applications. | To read more about BioMart, refer to the recent articles describing the BioMart software and its applications. | ||
Revision as of 05:16, 13 October 2011
| {{#icon: Biomart250.png|BioMart|200|BioMart}} {{#icon: October2011Logo.png|250px|center|October 2011 - Toronto |
2009 GMOD Summer School - Toronto, Canada}} | BioMart Session 2009 GMOD Summer School - Europe & |
__NOTITLE__
This tutorial walks you through how to install and configure a local installation of BioMart. This tutorial was originally taught by Junjun Zhang at the 2010 GMOD ??.
Contents
1. System overview and installation
Prerequisites for BioMart
Software: Java 1.6, Ant and SVN client
OS: major Linux distributions
Server: min. one GB memory, three GB for better performance
2. Downloading & Installing BioMart
2.1. Biomart 0.7 (MartJ -> MartBuilder + MartRunner)
Installation of BioMart has already been done on the VM image. Refer to BioMart Documentation for installation guidelines.
Download MartJ MartJ contains applications necessary to create a Mart: MarBuilder and MartRunner. Installation of MartJ has already been done on the VM image. Check Section 2 in Installing BioMart
Extract tarball content with:
$ # ! FYI only: this is already done for you on the VM ! $ mkdir ~/biomart_0_7 # arbitrary name of course $ mv ~/Downloads/martj-bin.tgz ~/biomart_0_7/ $ cd ~/biomart_0_7/ $ tar zxvf martj-bin.tgz # creates "martj" directory $ cd martj $ # ! FYI only: this is already done for you on the VM !
2.2. Biomart 0.8 (MartConfigurator)
Checkout latest release using SVN:
$ # ! FYI only: this is already done for you on the VM ! $ mkdir ~/biomart_0_8 # arbitrary name of course $ svn co https://code.oicr.on.ca/svn/biomart/biomart-java/branches/oct_3_2011 # creates "oct_3_2011" directory $ cd oct_3_2011 $ # build project with ant $ ant # in the future, you may use: ant clean dist $ # ! FYI only: this is already done for you on the VM !
3. Building mart
This section will show you how to create a (simplified) mart containing 1 dataset and based on the VEGA database from Ensembl, and using MartBuilder.
3.1. Describing mart (MartBuilder)
Creating/loading sample mart
3.1.1. Start MartBuilder
From the directory ~/biomart_0>7/martj, issue:
$ bin/martbuilder.sh
MartBuilder should open with an empty mart.
3.1.2. Add a schema
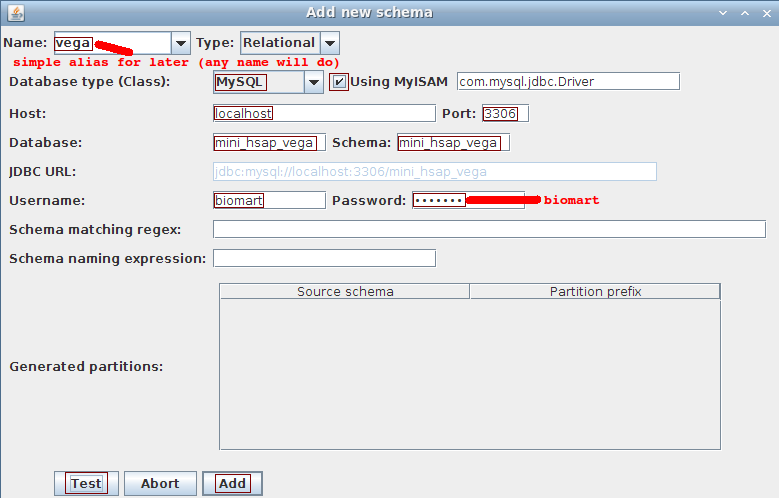
Choose Schema->Add to open the dialog to add a schema.
Fill in the fields using the following values:
- Name: vega
- Database type: MySQL
- Using MyISAM: checked
- Host: localhost
- Port: 3306
- Database: mini_hsap_vega
- Schema: mini_hsap_vega
- Username: biomart
- Password: biomart
ignore the last 3 fields (used for partitioning which is not covered in this tutorial)
Click the Test button to ensure we can connect to the database. Click the Add button in order to proceed with the dataset description.
You should now see the source database show in MartBuilder.
- You will see a blank mart, waiting to have one or more source schemata added to it and from which datasets will be generated later. The next step is to connect to an existing relational database and add the schema which contains the data which you wish to transform.
- 2. Choose Add from the Schema menu. Input connection parameters as follows:
- The proper information for the Target database and Target schema fields
- Note that if you did not specify a schema when creating your database, your tables will be in the default schema for your platform:
- MySQL: does not have schema, will be the same as the database name
- PostGreSQL: “public”
- Oracle and DB2: the username of the user who created the database
- SQL Server: “dbo”
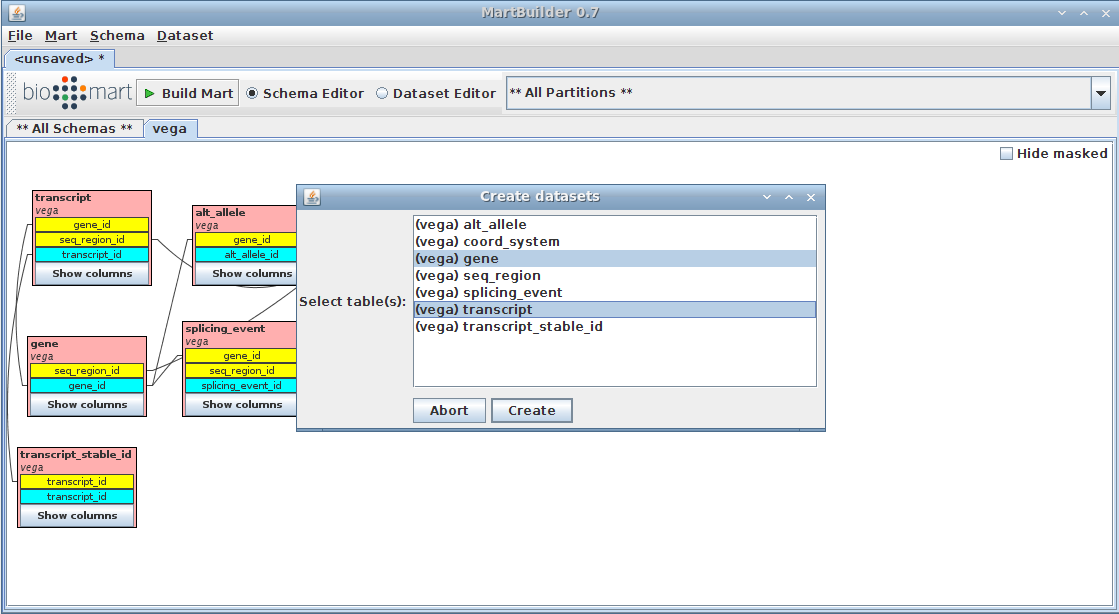
3.1.2. Choose dataset
We are going to create a dataset based on the tables "gene" and "transcript" (as main and submain tables respectively, as described in presentation)
Fill in the fields using the following values:
- Right-click on the "gene" table
- Click on Create dataset TODO
the gene table should be highlighted already as we arrived on the current menu by clicking on it
- add the transcript table (standard "Ctrl + click)
- Press the Create button
- 3. Create datasets
Now you should be able to see the VEGA schema.
Find the table that contains the data you wish to use in the main table(s) of your dataset. Right-click on the table, and choose Create dataset. Here we selected both Gene and Transcript tables to be the main tables.
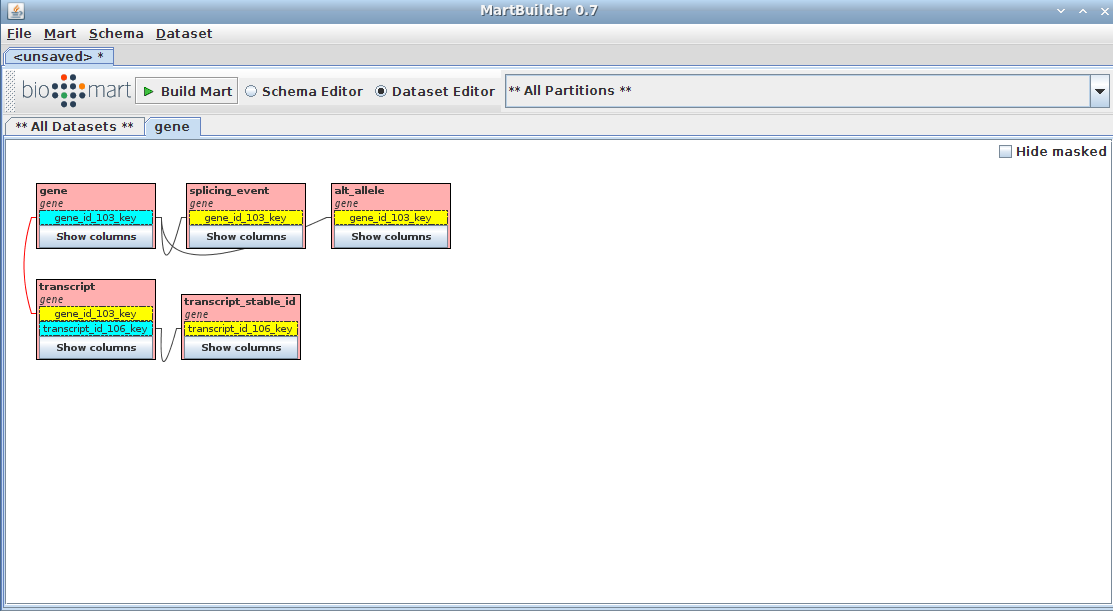
Click Create. Dataset will look like this:
3.2 Materializing mart (MartBuilder->MartRunner)
Your dataset does not actually exist yet. In order to create it, you need to generate the SQL for it then run that SQL against your database. BioMart offers a tool (MartRunner) that does that for you, using JDBC. The SQL used is as ANSI-compliant as possible, with some exceptions based on the RDBMS in use.
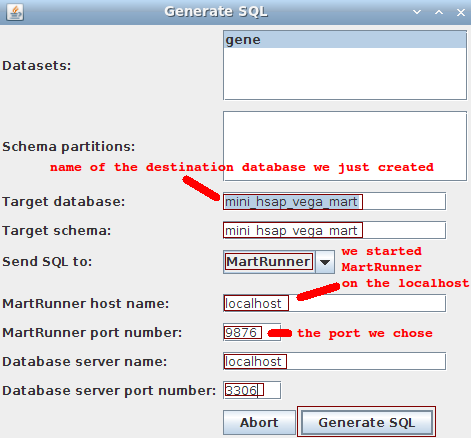
4. We now going to transform the source data into target dataset, but before that, we have to create a target database:
$ MY_DESTINATION_DATABASE=mini_hsap_vega_mart $ mysql -hlocalhost -P3306 -ubiomart -pbiomart -e "create database $MY_DESTINATION_DATABASE"
Also we have to have martrunner running. Lets run it over a local port (any free one will do).
5. Start martrunner with:
$ MY_MART_RUNNER_PORT=9876 $ bin/martrunner.sh $MY_MART_RUNNER_PORT
MartBuilder will send the materialization SQL to MartRunner through that port (in this example, 9876), and MartRunner will execute the transformation SQL. Note that MartRunner may be run from another machine, as we specify its connection parameters to MartBuilder (see below).
We go back to MartBuilder, and click on the BuildMart button to pop up the following dialog:
The proper information for the Target database and Target schema fields differ depending on the database server type:
MySQL-- Target database and target schema must be the same, and different than the original source database and schema. The database must exist on the server.
SQL Server -- Target database and target schema both must exist on the server. The original source schema should not be used.
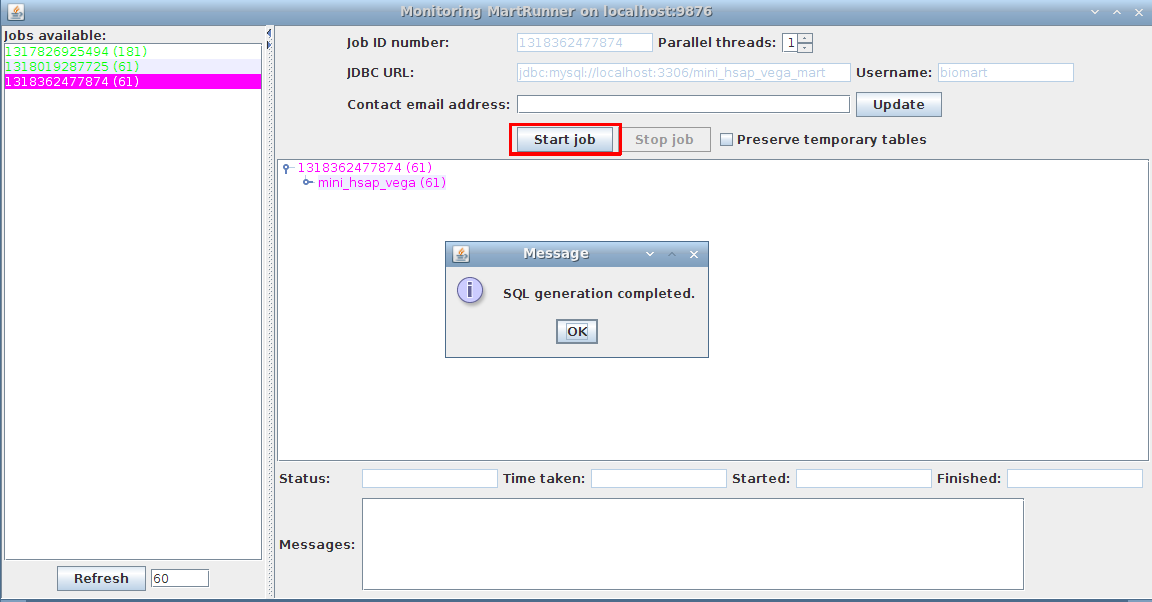
PostgreSQL, Oracle, and DB2 -- Target database must be the same as the original source. The target schema should exist within this database, and should be different than the original source schema. Start job in MartRunner (windows should have poped up upon SQL generation):
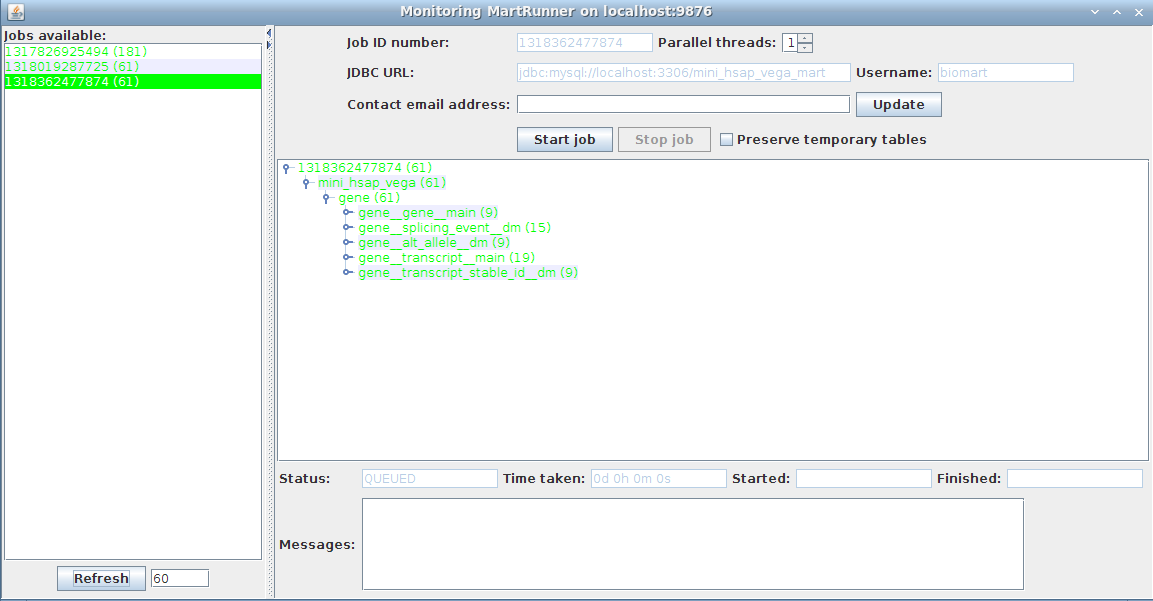
5. Monitor progress using the Monitor MartRunner progress
Ensure everything is successful (green font):
Your schema now contains a mart with a complete dataset ready for configuring with MartConfigurator.
4. Configuring mart (MartConfigurator)
This section will show you how to configure the created VEGA mart using MartConfigurator
4.1 Run MartConfigurator
Run MartConfigurator with the following command in the directory of your installation:
$ dist/scripts/martconfigurator.sh
Add Source
In the MartConfigurator window, click *Add Mart *in the upper right corner.
A list of available marts will appear in the left corner. Click mini_hsap_vega_mart, and click Add. The new mart will appear in the left-hand corner of the MartConfigurator window.
VIEW ??? Schema, management window??
Create and configure access point.
To create an access point click on the +Add Access Point button in the right-hand Portal section.
You will be given a list of the existing marts to choose which one you would like to make an access point for. After giving the new access point a name of your choice, it will appear in the GUI tab. Double-clicking on the access point will open a new window that allows you to modify the access point.
The display name of any object (a container, attribute, or filter) can be changed by selecting that object (by clicking on it) and then double clicking the displayname property in the lower right-hand pane.
Deploy BioMart Server
To test the registry, you can now click on the Start Server button in the upper right. You will be prompted to save the registry file you have created, and after doing so, the server will be deployed on your local machine, port 9000. Your web page should open automatically when the local server is ready. You can also deploy BioMart from the command line on the server. To deploy BioMart, from the directory of your installation, run the following command:
./dist/scripts/biomart-server.sh start
To Stop the server, use the command:
./dist/scripts/biomart-server.sh stop More exercises with MartConfigurator
Set Filter typesSet Attribute URLSelect GUI type
Creating Links between sources
If two data sources contain common information (e.g. a Gene/Protein ID), this can be used to create a link, allowing filters and attributes from one data source to appear in the other. These are called “pointer attributes” and “pointer filters,” and the attribute or filter to which they point is called the “target. ”To add a pointer to an access point, double click on that access point in the portal tab to edit it. In the top left corner of the editing window, click on the Import from sources button.
You will be given a list of the existing data sources to choose which one you would like to make an access point for. After giving the new access point a name of your choice, it will appear in the GUI tab. Double-clicking on the access point icon will open a new window that allows you to modify the access point. References
5. Querying a BioMart server via REST API
5.1 MetaData queries
5.2 Data query
To read more about BioMart, refer to the recent articles describing the BioMart software and its applications.
1. Zhang J, Haider S, Baran J, Cros A, Guberman JM, Hsu J, Liang Y, Yao L, Kasprzyk A. BioMart: a data federation framework for large collaborative projects. Database (Oxford). 2011 Sep 19:2011:bar038.
2.Guberman JM, Ai J, Baran J., et al. BioMart Central Portal: An Open Database Network for the Biological Community. Database (Oxford). 2011 Sep18;2011:bar041.
3. Zhang J, Baran J, Guberman JM, Haider, S, Hsu J, Liang Y, Rivkin E, Wang J, Whitty B, Wong-Erasmus M, Yao L, Kasprzyk A. International Cancer Genome Consortium Data Portal - a One-stop Shop for Cancer Genomics Data. Database (Oxford). 2011 Sep19;2011:bar026.