Difference between revisions of "BioMart Tutorial 2011"
Anthony Cros (Talk | contribs) (→Description) |
Anthony Cros (Talk | contribs) (→Importing attribute) |
||
| Line 465: | Line 465: | ||
# Boolean filter list: no covered here | # Boolean filter list: no covered here | ||
== Importing attribute == | == Importing attribute == | ||
| − | # We want to add a pathway attribute to the vega "Form" GUI (of MartForm type). In order to do that we must: | + | #: We want to add a pathway attribute to the vega "Form" GUI (of MartForm type). In order to do that we must: |
| − | + | # Under the "Form" GUI tab, double-click on the "gene_vega_ap" access point to open configuration dialog for it | |
| − | + | #: It is important to open "gene_vega_ap" first as opposed to "pathway_ap", since we are going to add a "pathway" attribute to "gene_vega" and not the opposite | |
| − | + | # Use the ''Show both'' view | |
| − | + | # On the right-hand side (''Access Point'' view), select the "root --> Attributes" container | |
| − | + | # Right-click --> ''Add container'' | |
| − | + | # Type in name, for instance "Pathway" since we're importing a pathway attribute | |
| − | + | # On the left-hand side (''Source'' view), change source to ''pathway'' | |
| − | + | # In the ''Source'' view, search for the filter called "Pathway name" (Using the ''Find'' button) | |
| − | + | # Drap-and-drop the selected attribute on the left-hand side ("Pathway name") into the newly created container in the right-hand side ("Pathway") | |
| − | + | #: Note: this will open the link dialog as we need to specify how to link those 2 datasets | |
| − | + | # You can keep default link name ("pathway-gene_vega-link") and click ''Next'' | |
| − | + | # Choose attribute(s) to join on (this should not be the same attribute as the one we are importing as it would not add any information, although nothing prevents it) | |
| − | + | #* left-hand side: In the ''Source'' view, search for the filter called "hsgene" (Using the ''Find'' button) | |
| − | + | #* Double-click on it to add it | |
| − | + | #* right-hand side: In the ''Access Point'' view, search for the filter called "Gene ENSEMBL ID" (Using the ''Find'' button) | |
| − | + | #* Double-click on it to add it | |
| − | + | #: Note that one could link on more than one attribute provided that there are an equal number of attributes on each side | |
| − | + | #: [[File:Mc_link.png]] | |
| − | + | # Keep default setting for the next page and ''Finish'' | |
| − | + | # The newly added attribute should now show in the gene_vega_ap access point | |
# Re-deploy the server and observe behavior of the imported pathway attribute | # Re-deploy the server and observe behavior of the imported pathway attribute | ||
| − | + | ||
== Importing filter == | == Importing filter == | ||
# Follow the same steps as with "Pathway name" attribute for its counterpart filter ("root --> Filters --> FILTERS: --> FEATURES: --> Pathway --> Pathway name") | # Follow the same steps as with "Pathway name" attribute for its counterpart filter ("root --> Filters --> FILTERS: --> FEATURES: --> Pathway --> Pathway name") | ||
Revision as of 22:52, 13 October 2011
TODO: add small screenshot to exercises?
TODO: REST?
TODO: get svn revision
TODO: recreate image
TODO: upload image
TODO: test each commands again
| {{#icon: Biomart250.png|BioMart|200|BioMart}} {{#icon: 170px-October2011Logo.png|October 2011 - Toronto |
2009 GMOD Summer School - Toronto, Canada}} | BioMart Session 2011 GMOD Community Meeting & |
__NOTITLE__
This tutorial walks you through how to install and configure a local installation of BioMart.
Contents
System overview and installation
Prerequisites for BioMart:
- Software: Java 1.6, Ant and SVN client
- OS: Linux, Mac & Windows
- Server: min. 1 GB memory, 3 GB for better performance
Accounts
- linux:
- username: "biomart" (sudoer, for instance: "sudo echo hello" runs echo command as root)
- pasword: "biomart"
- mysql:
- username: "biomart"
- pasword: "biomart"
Virtual machine
- Please download the image file (see OVF/OVA format) from www.biomart.org/lubuntu.ova (TODO: get final location) or ask for the USB memory stick to one of our merry instructors
- If you have not already done so, please download VirtualBox (prefered over VMware since its free) and follow the installation instructions for your platform.
- Once VirtualBox is installed and you have downloaded the image file (.ova):
- Start VirtualBox with:
- $ virtualbox
- Import image with: (from the manual menu) Machine --> Import TODO: exact menu name
- Start VirtualBox with:
Downloading & Installing BioMart
Two components are necessary for this tutorial: MartBuilder is installed form an older version of BioMart (0.7), and MartConfigurator is installed from the most recent version (0.8) (until all features are fully ported to the new version)
Installation of MartBuilder and MartConfigurator has already been done on the VM image under ~/biomart_0_7.template and ~/biomart_0_8.template, but we are going to do once together for demonstration purposes.
Refer to BioMart Documentation for more information.
Biomart 0.7 (MartJ -> MartBuilder + MartRunner)
MartJ contains applications necessary to create a Mart: MarBuilder and MartRunner.
Check Section 2 in Installing BioMart for more information.
Download & extract tarball content with:
$ mkdir ~/biomart_0_7 # arbitrary name of course $ cd ~/biomart_0_7 $ wget ftp://anonymous@ftp.ebi.ac.uk/pub/software/biomart/martj_current/martj-bin.tgz # or get it from MartJ $ tar zxvf martj-bin.tgz # creates "martj" directory $ cd martj $ ls bin/martbuilder.sh bin/martrunner.sh # what we care about
Biomart 0.8 (MartConfigurator)
Checkout latest release using SVN:
$ mkdir ~/biomart_0_8 # arbitrary name of course $ cd ~/biomart_0_8 $ svn co -r 32343 https://code.oicr.on.ca/svn/biomart/biomart-java/branches/oct_3_2011 martconfigurator # creates "martconfigurator" directory $ cd martconfigurator $ ant # build project with ant; in the future, you may use: ant clean dist $ ls dist/scripts/martconfigurator.sh dist/scripts/biomart-server.sh # what we care about
Building mart
This section will show you how to create a (simplified) mart containing 1 dataset based on the VEGA database, using MartBuilder.
Creating mart schema (MartBuilder)
Start MartBuilder
From the directory ~/biomart_0_7/martj, issue:
$ bin/martbuilder.sh
MartBuilder should open with an empty mart.
Add a source schema
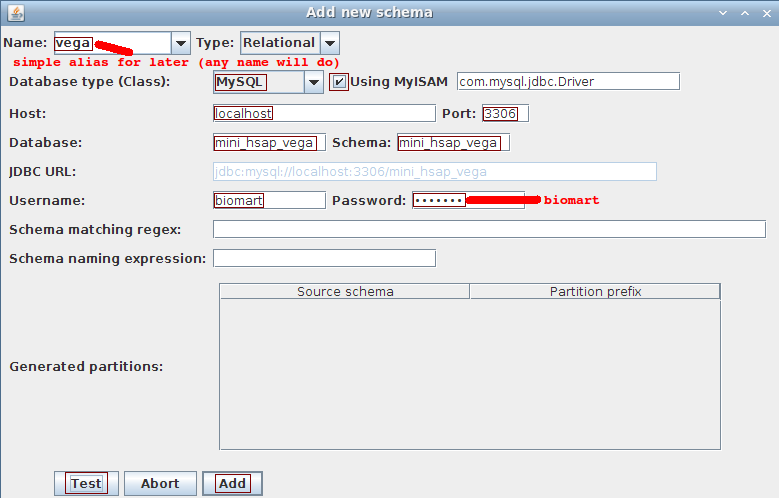
In the manual bar, choose Schema->Add to open the dialog to add a schema.
- Name: vega
- Database type: MySQL
- Using MyISAM: checked
- Host: localhost
- Port: 3306
- Database: mini_hsap_vega
- Schema: mini_hsap_vega
- Username: biomart
- Password: biomart
ignore the last 3 fields (used for partitioning which is not covered in this tutorial)
Click the Test button to ensure we can connect to the database. Click the Add button in order to proceed with the dataset description.
You should now see the source database shown in MartBuilder (see screenshot in the next section).
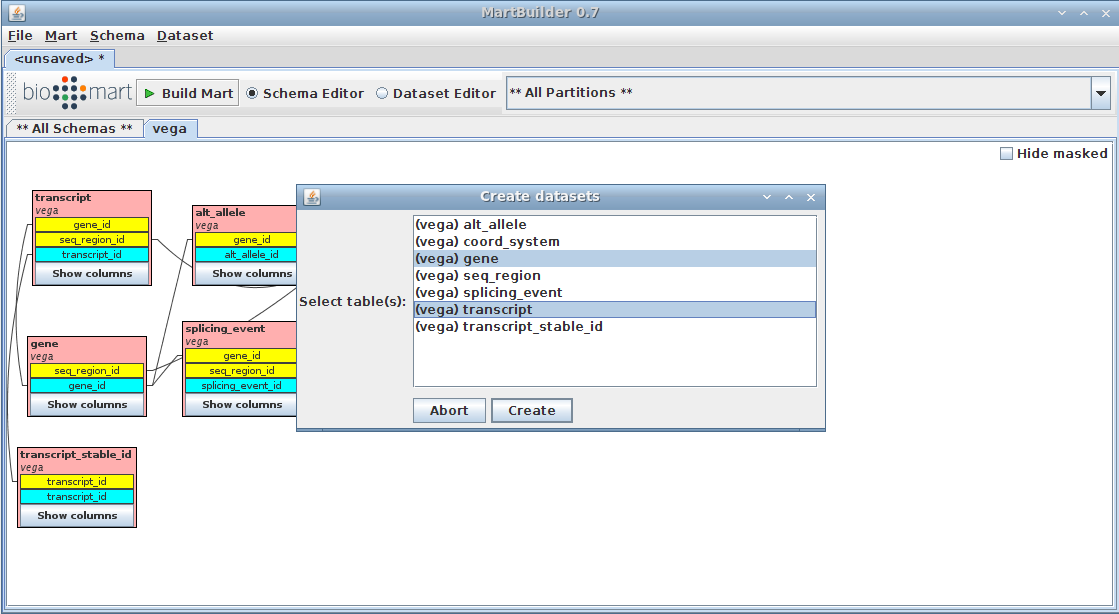
Select main table(s)
We are going to create a dataset based on the tables "gene" and "transcript" (as main and submain tables respectively, as described in presentation)
- Right-click on the "gene" table
- Click on Create dataset
the gene table should be highlighted already as we arrived on the current menu by clicking on it
- add the transcript table (standard "Ctrl + click)
- Press the Create button
Materializing mart (MartBuilder->MartRunner)
Your dataset does not actually exist yet. In order to create it, you need to generate the SQL for it then run that SQL against your database.
BioMart offers a tool (MartRunner) that does that for you, using JDBC. The SQL used is as ANSI-compliant as possible, with some exceptions based on the RDBMS in use.
1. We now going to transform the source data into target dataset, but before that, we have to create a target database:
$ mysql -hlocalhost -P3306 -ubiomart -pbiomart -e "create database mini_hsap_vega_mart"
In theory one could materialize a mart in the same database as the source one provided there is no table name conflicts, but this is strongly discouraged.
Also we have to have MartRunner running. Lets run it over a local port (any free one will do).
2. Start martrunner with:
$ bin/martrunner.sh 9876
MartBuilder will send the materialization SQL to MartRunner through that port (in this example, 9876), and MartRunner will execute the transformation SQL.
Note that MartRunner may be run from another machine, as we specify its connection parameters to MartBuilder (see below).
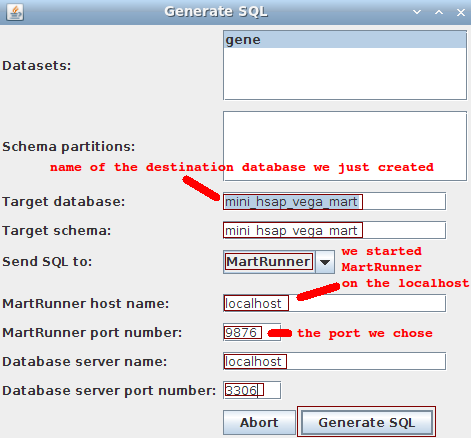
We go back to MartBuilder, and click on the Build Mart button to pop up the following dialog:
- Datasets: gene
- Schema partitions: ignore
- Target database: mini_hsap_vega_mart (the one we just manually created)
- Target schema: mini_hsap_vega_mart
- Send SQL to: MartRunner (other options are available)
- MartRunner host name: localhost
- MartRunner port number: 9876 (the one we just arbitrarily chose because it was free)
- Database server name: localhost
- Database server port number: 3306
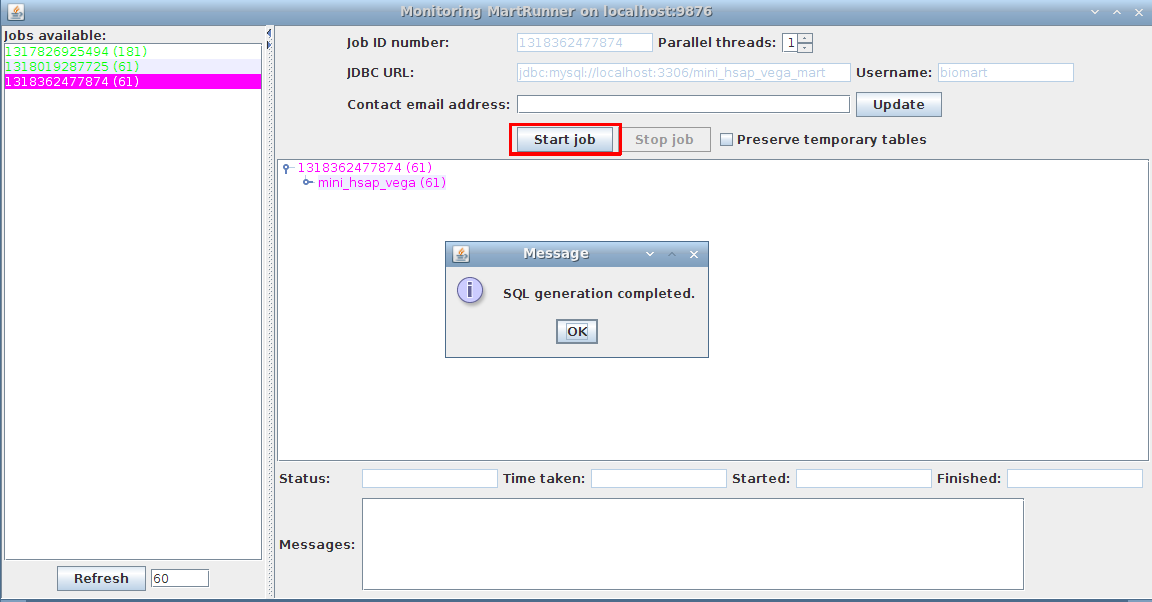
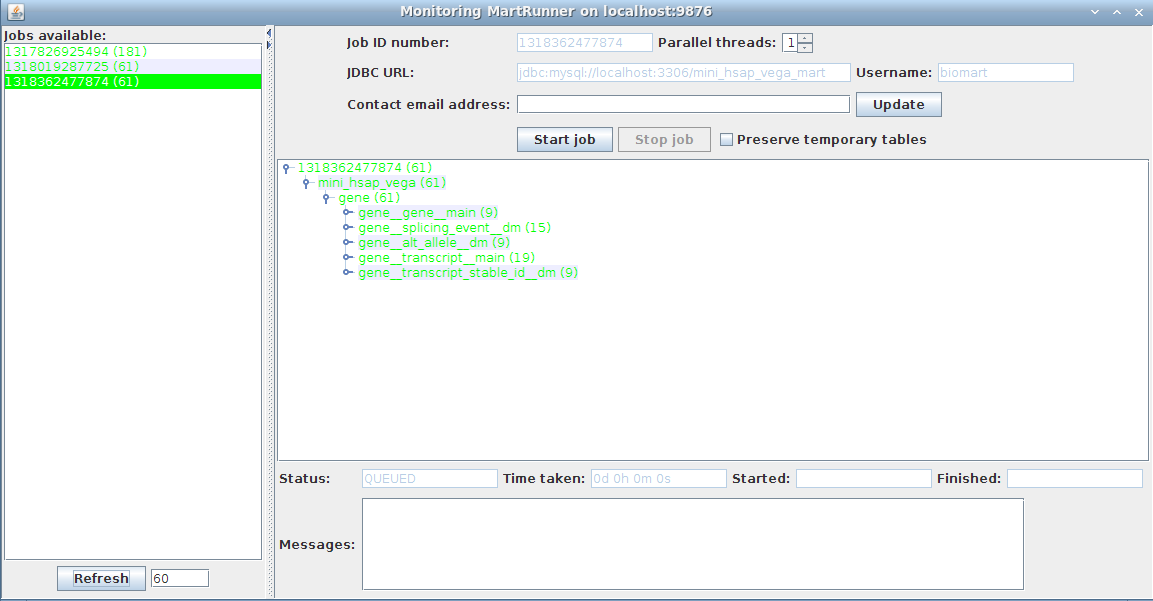
3. Monitor progress using the Monitor MartRunner progress
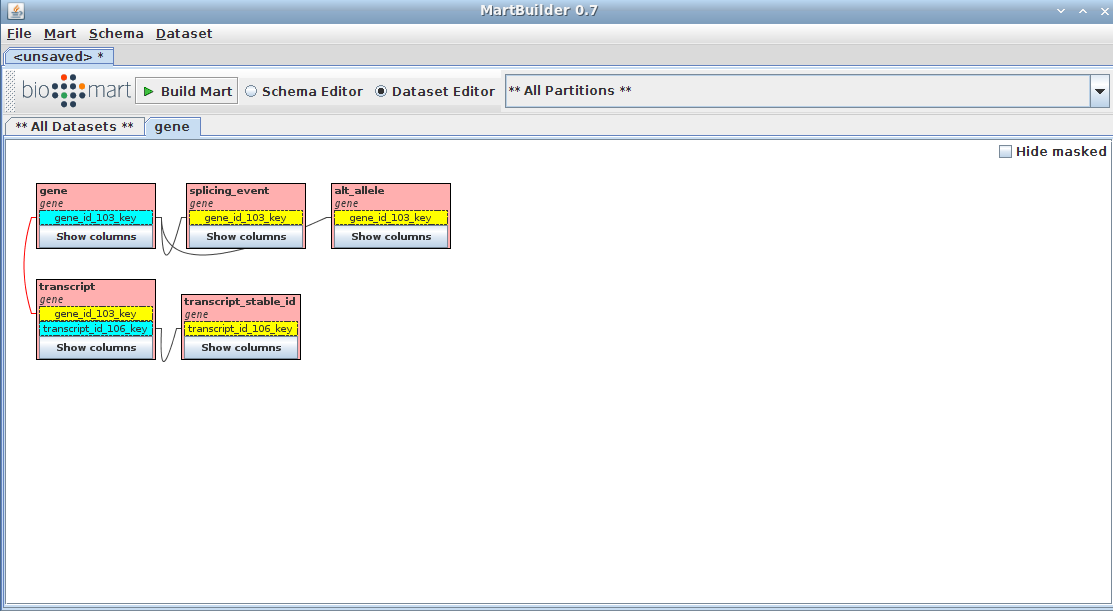
Your schema now contains a mart with a complete dataset ready for configuring with MartConfigurator.
Configuring mart (MartConfigurator) - basic
This section will show you how to configure the created VEGA mart using MartConfigurator
Start MartConfigurator
Start MartConfigurator with the following command in the directory of your installation:
$ cd ~/biomart_0_8/martconfigurator $ dist/scripts/martconfigurator.sh
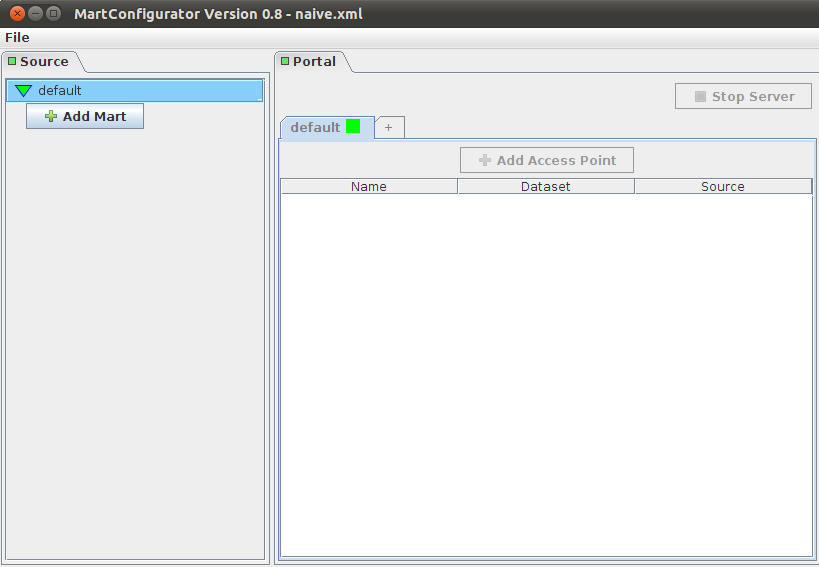
The panel on the left corresponds to datasources; in our case we only have one mart that has one dataset: mini_hsap_vega_mini, but there could be more
The panel on the right corresponds to access points for those datasources
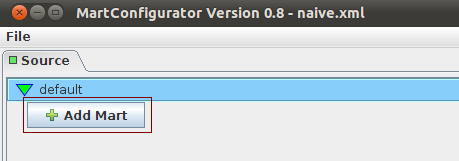
Add Mart
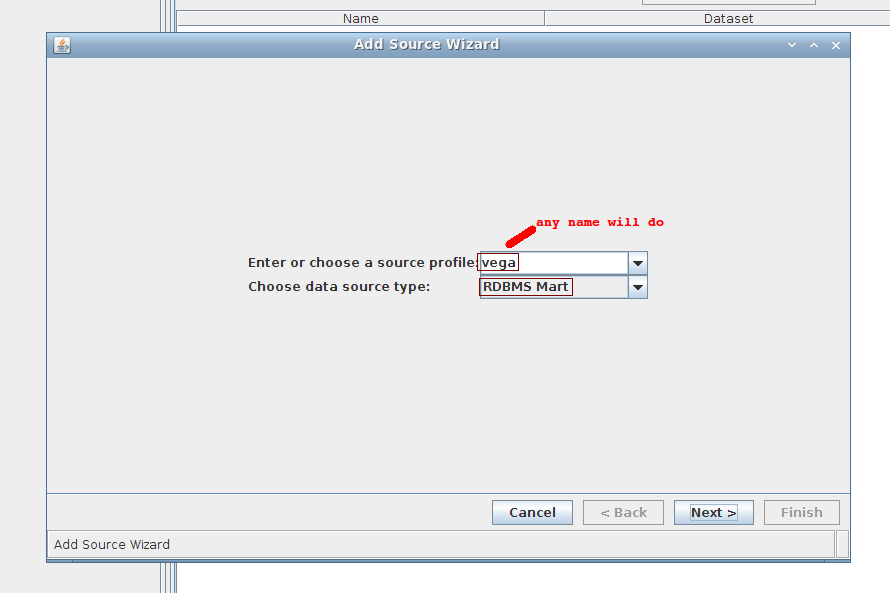
1. Wizard step 1/4:
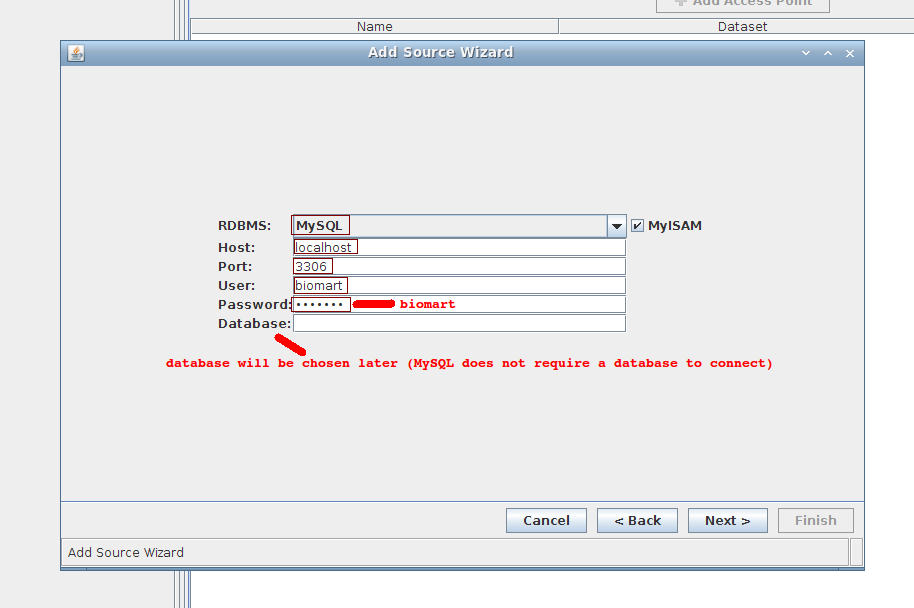
2. Wizard step 2/4:
- RDBMS: MySQL (keep MyISAM)
- Host: localhost
- Port: 3306
- User: biomart
- Password: biomart
- Database: can leave empty for now
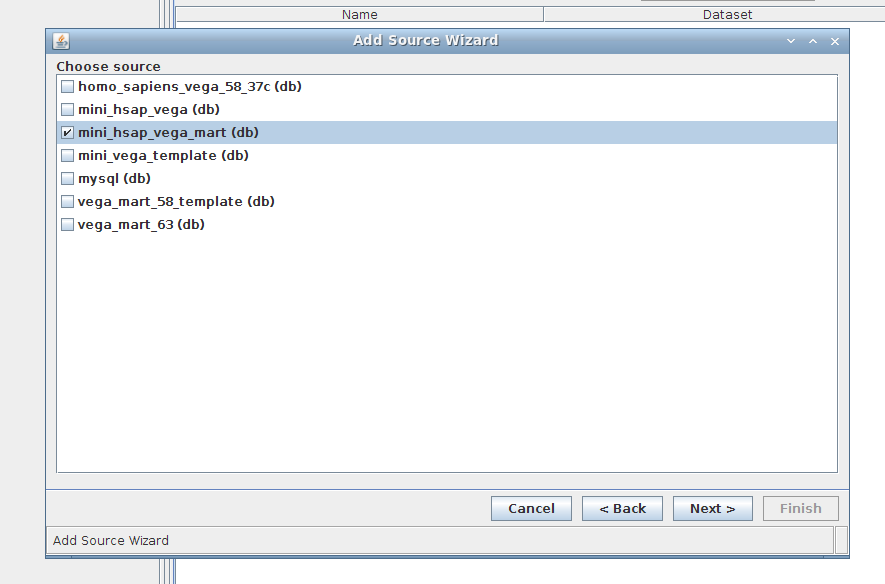
3. Wizard step 3/4:
Select mini_hsap_vega_mart, the mart that we just built using MartBuilder/MartRunner and based upon the mini_hsap_vega database
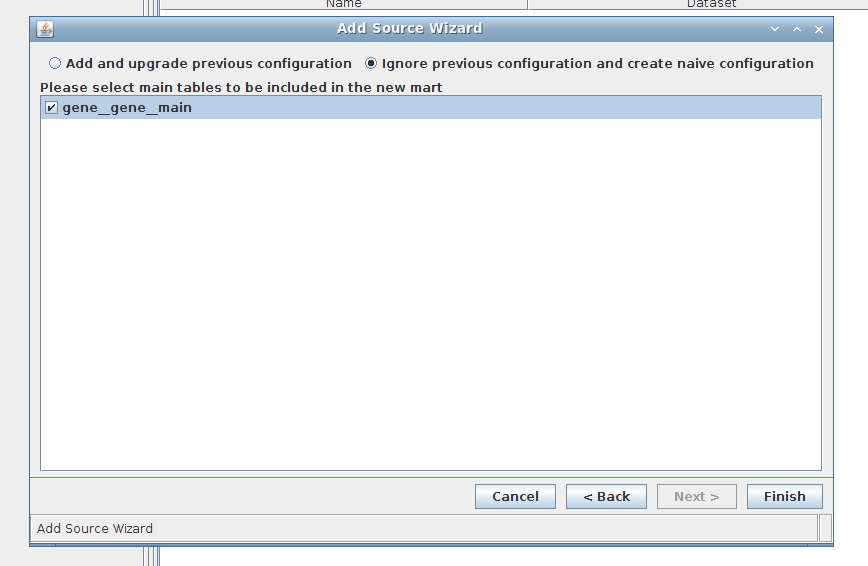
4. Wizard step 4/4:
- Create naive configuration
- Choose the main table "gene__gene__main" (only one) TODO: has this changed?
5. Done!
Creating Access Point
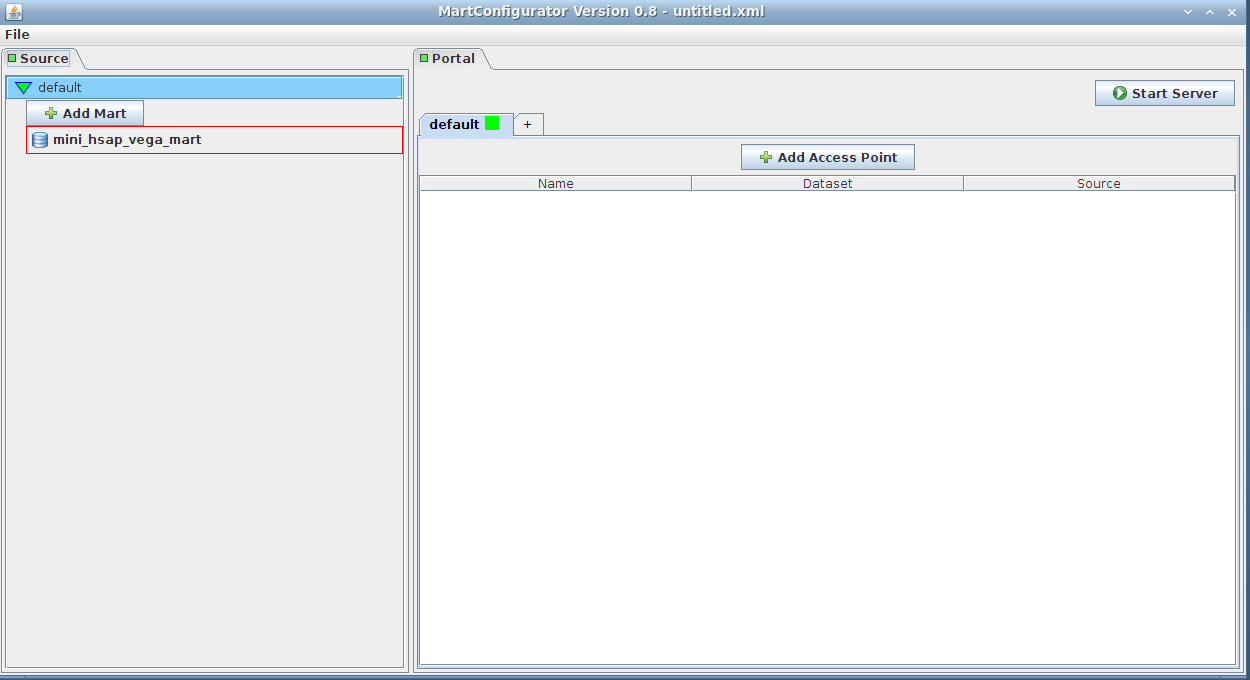
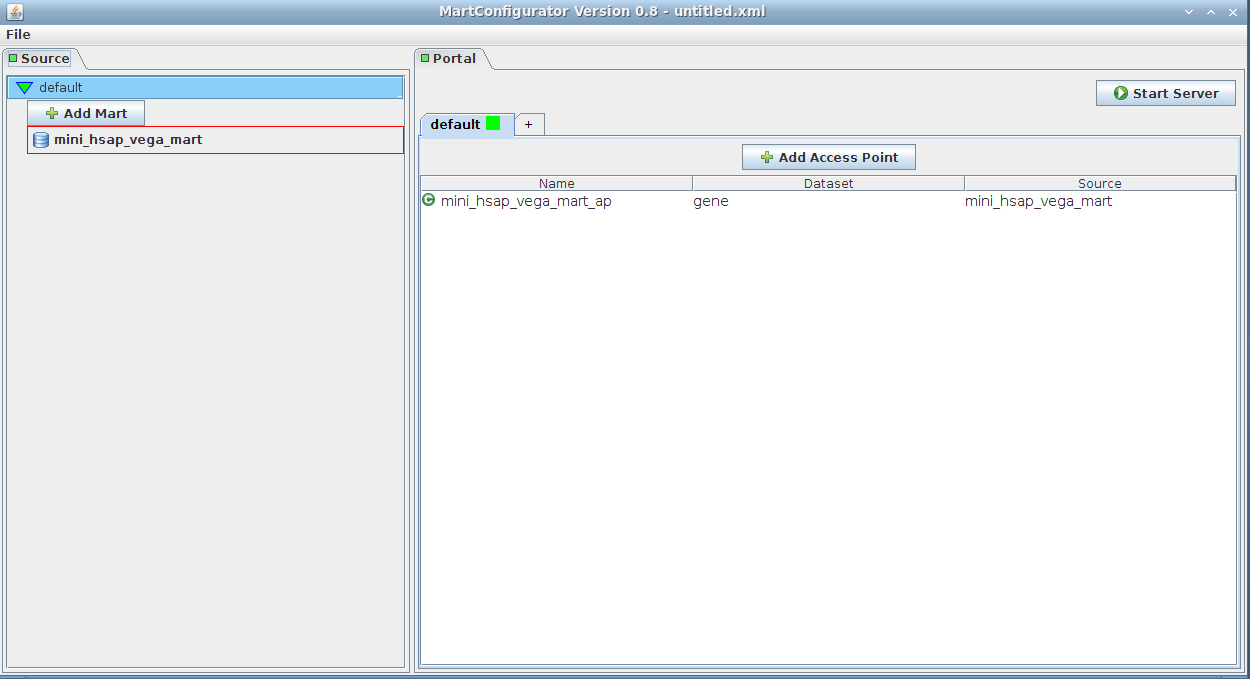
Simply drag-and-drop the source from the left side (Source frame) to anywhere on the right side (Portal frame)
It will add an access point to the mart. The default name is mini_hsap_vega_mart_ap, ap standing for Access Point, but you can give it a name of your choice.
Note that clicking the Add Access Point button would yield the same result. You would then be given a list of the existing marts to choose from.
The drag-and-drop access point will contain everything, as opposed to the one created with the Add Access Point button, which by default adds an empty (blank) access point. The later can be changed to contain everything as well by unchecking the blank checkbox
Deploying BioMart Server (Jetty)
In order to deploy the application on Jetty, one has 2 options:
- Use the Start Server button in the upper right corner
- This will result in prompting a dialog for saving the current state - referred to as registry
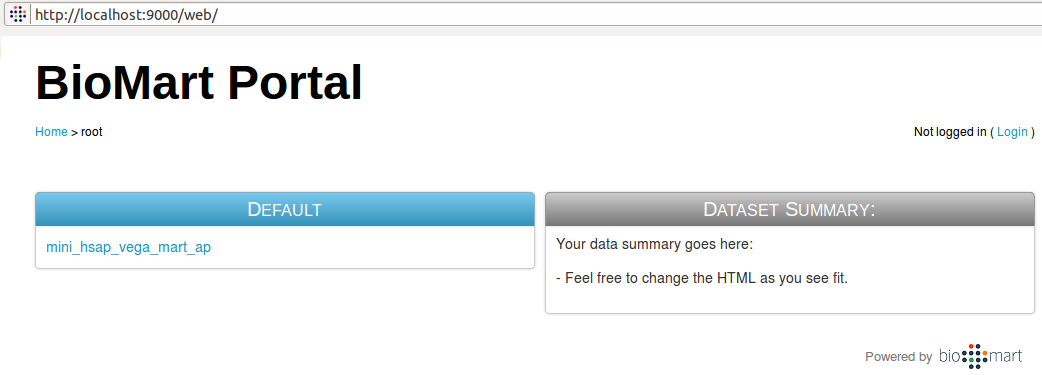
- The server will be deployed on your local machine, port 9000 by default (unless specified differently in .biomart.properties file).
- Your web page should open automatically when the local server is ready.
Stopping the server
- Use the Stop Server button in the upper right corner
Alternative way to deploy
There is also a command-line approch to starting/stopping the server using:
$ dist/scripts/biomart-server.sh start $ dist/scripts/biomart-server.sh stop
but that will not be covered here (involves the registry in a specific directory first)
Using deployed application
The interface is intuitive, typical interaction involve choose a number of attributes and filters and obtaining results accordingly.
Configuring mart (MartConfigurator) - more advanced
From the configuration editor (see previous section)
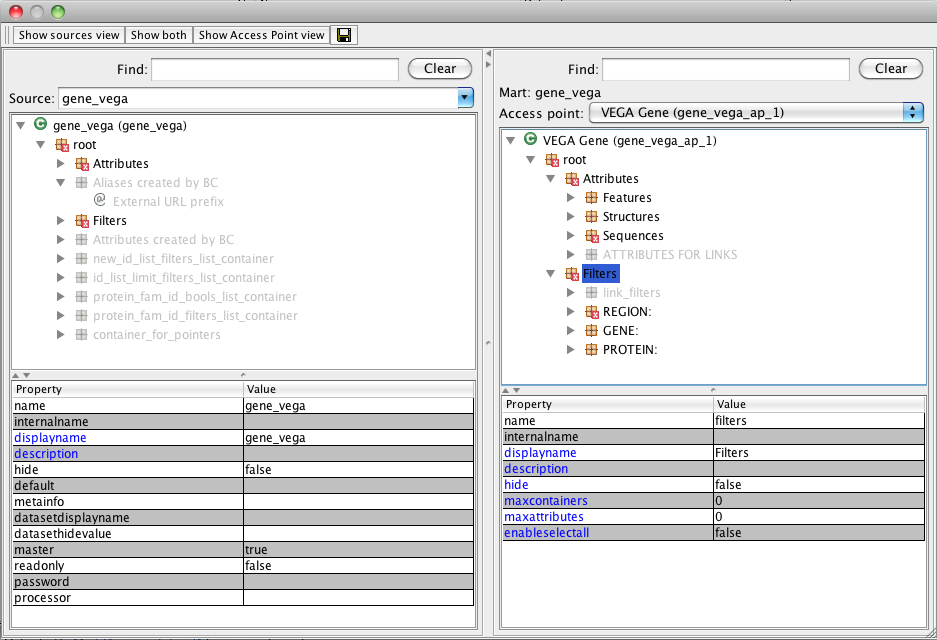
Configuration editor
Description
- top panel: Configuration tree
- Show sources view: where one can modify config in a way that will affect all access points
- Show access point view: where one can modify individual access point's configs specifically
- Show both: most typical usage
- top-left panel: source view
- top-right panel: access point view
- bottom panel: Editor frame
- bottom-left panel: property names
- bottom-right panel: property values
Notes
Only properties whose name appears in blue font can be modified.
Each view has a Find search box that allows to quickly find attributes/filters based on their names
Also very convenient, the attributes and filters in each view offer an Show in the [opposite view] item in their context menu. It filters the opposite view side to show the counterpart filter/attribute
TODO merge whole section to exercices below == Containers == Typical properties that can be changed: * displayname * description * hide: whether attribute should be hidden or not * enableselectall: offer checkbox allowing selection of all attributes/filters in the given container == Attributes == Typical properties that can be changed: * displayname * description * hide * linkouturl: to make result values linkable to an external resource * datatype: String, Integer, Float, Boolean (useful mostly for reordering) == Filters == Typical properties that can be changed: * displayname * description * type (covered in the next section) * qualifier: whether to use "=", "<=", ">=", "LIKE" operations * spliton: separator for filter list values (see below) == Filters types == === Regular filters === Typically used to (suprisingly) filter data based on values of interest (for instance a specific gene, or location, or type, ...) === Filter lists === They are considered filter themselves, but actually contain a list of regular filters === Linking === Set Filter typesSet Attribute URLSelect GUI type Creating Links between sources If two data sources contain common information (e.g. a Gene/Protein ID), this can be used to create a link, allowing filters and attributes from one data source to appear in the other. These are called “pointer attributes” and “pointer filters,” and the attribute or filter to which they point is called the “target. ”To add a pointer to an access point, double click on that access point in the portal tab to edit it. In the top left corner of the editing window, click on the Import from sources button. You will be given a list of the existing data sources to choose which one you would like to make an access point for. After giving the new access point a name of your choice, it will appear in the GUI tab. Double-clicking on the access point icon will open a new window that allows you to modify the access point. References
More exercises with MartConfigurator
Rename 'default' GUI tab to "Form"
- TODO: explain GUI tab
- Right-click on the tab --> Rename
- enter new name: "Form"
Add new GUI tab
- Click on the "+" sign next to the latest GUI tab
- Enter name of the new tab: "Wizard"
- Right-click on the newly created tab --> Set GUI type --> MartWizard
Add a URL-based marts source
- Add remote mart (URL Mart) Using backward compatibility (from previous BioMart versions: <= 0.7)
- In the Source view: Add Mart
- Select URL Mart --> Next
- Input the following values:
- Protocol: http
- Host: www.biomart.org
- Port: 80
- Path: /biomart/martservice
- Choose source: "vega (url)" --> Next (we will only need "REACTOME" later)
- Select all 3 datasets and click Next
- Uncheck import each datasets to individual marts, one dataset per mart --> Finish
- Backward compatibility is run in the background in order to convert a mart configuration in 0.7 format to one in 0.8
- A datasource called "gene_vega" should appear on the right-hand side panel (datasource panel)
- Reproduce the same process for "REACTOME (url)", we will need it later
Add access points for the URL marts
- For both "vega" and "REACTOME", add access points to the "Form" GUI tab
- For both "vega" and "REACTOME", add access points to the "Wizard" GUI tab
Re-deploy server
- observe changes
Attributes
- Attribute linkouturl:
- TODO: example
- Attribute datatype:
- TODO: example
Filters
- Single select: "Chromosome"
- Open the configuration panel and choose Show both view
- In the Source view, search for the filter called "Chromosome" (Using the Find button)
- Change the type to singleSelect in the bottom-left panel (property editor for the Source view)
- Multi-select: "Gene type"
- In the Source view, search for the filter called "Gene type" (Using the Find button)
- Change the type to multiSelect in the bottom-left panel (property editor for the Source view)
- Dropdown options: "Chromosome" & "Gene type"
- Right-click on the filter of interest ("Chromosome" & "Gene type")
- Select Dropdown options in the menu
- For each dataset (hsapiens, mmusculus, drerio), click on the Update button in order to populate the values based on the database content
- Note that one may also manually add/remove values
- Composite filter: "Multiple Chromosomal Regions"
- In the Source view, search for the filter called "Multiple Chromosomal Regions (Chr:Start:End:Strand)" (Using the Find button)
- Change the following properties:
- type: upload (temporary name - under development)
- spliton: ":" (filter values are separated by a colon - do not include the quotes obviously)
- operation: and (only one available for now)
- filterlist: chromosome_name,start,end,strand (the 4 regular filters composing this composite filter)
- Boolean filter list: no covered here
Importing attribute
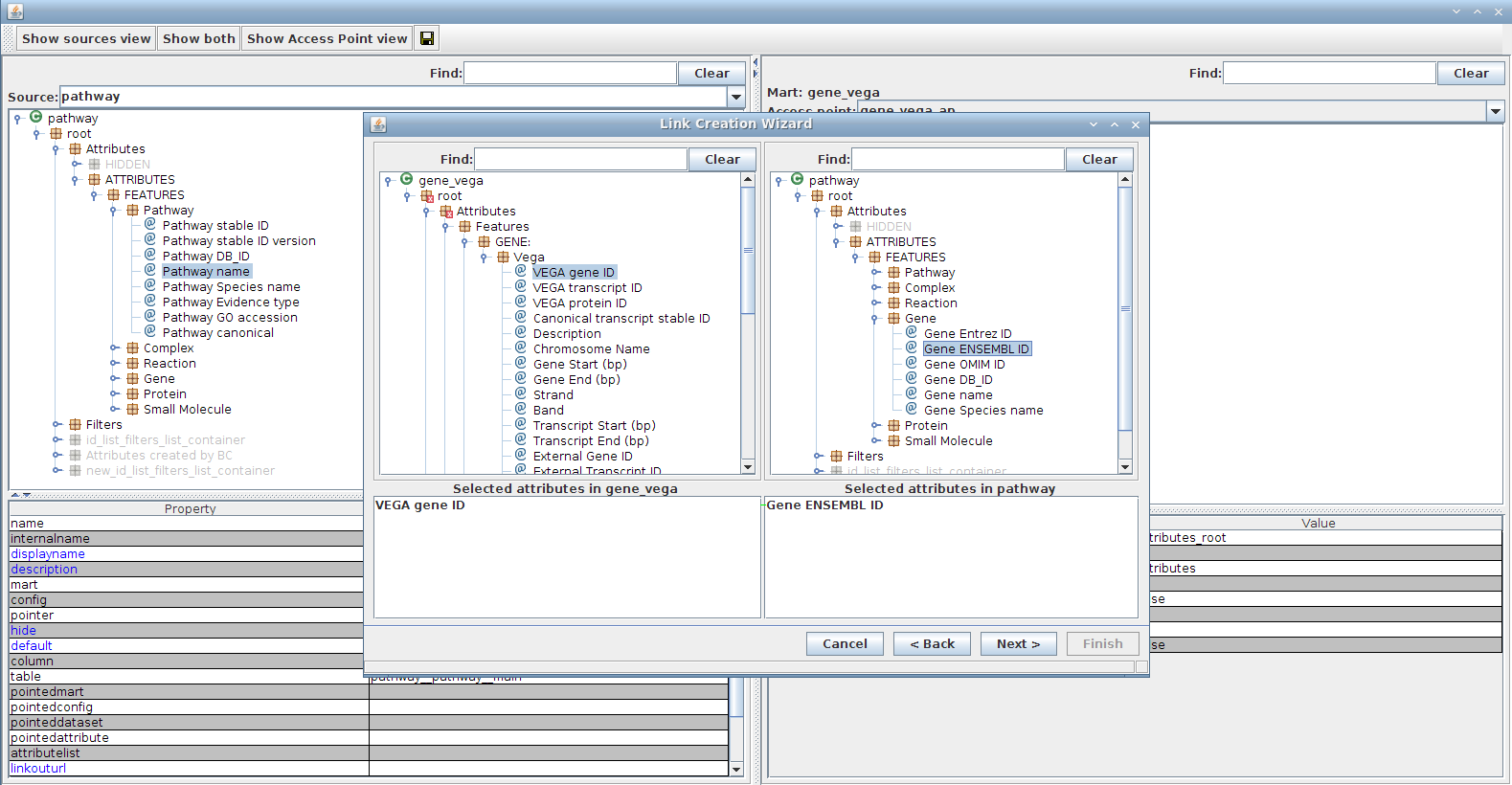
- We want to add a pathway attribute to the vega "Form" GUI (of MartForm type). In order to do that we must:
- Under the "Form" GUI tab, double-click on the "gene_vega_ap" access point to open configuration dialog for it
- It is important to open "gene_vega_ap" first as opposed to "pathway_ap", since we are going to add a "pathway" attribute to "gene_vega" and not the opposite
- Use the Show both view
- On the right-hand side (Access Point view), select the "root --> Attributes" container
- Right-click --> Add container
- Type in name, for instance "Pathway" since we're importing a pathway attribute
- On the left-hand side (Source view), change source to pathway
- In the Source view, search for the filter called "Pathway name" (Using the Find button)
- Drap-and-drop the selected attribute on the left-hand side ("Pathway name") into the newly created container in the right-hand side ("Pathway")
- Note: this will open the link dialog as we need to specify how to link those 2 datasets
- You can keep default link name ("pathway-gene_vega-link") and click Next
- Choose attribute(s) to join on (this should not be the same attribute as the one we are importing as it would not add any information, although nothing prevents it)
- left-hand side: In the Source view, search for the filter called "hsgene" (Using the Find button)
- Double-click on it to add it
- right-hand side: In the Access Point view, search for the filter called "Gene ENSEMBL ID" (Using the Find button)
- Double-click on it to add it
- Keep default setting for the next page and Finish
- The newly added attribute should now show in the gene_vega_ap access point
- Re-deploy the server and observe behavior of the imported pathway attribute
Importing filter
- Follow the same steps as with "Pathway name" attribute for its counterpart filter ("root --> Filters --> FILTERS: --> FEATURES: --> Pathway --> Pathway name")
- The link already exists so there will be no link creation dialog opening
- Re-deploy the server and observe behavior of the imported pathway filter
Querying a BioMart server via REST API
MetaData queries
http://localhost:9000/martservice/marts
http://localhost:9000/martservice/datasets?mart=gene_vega
http://localhost:9000/martservice/accesspoints?datasets=&mart=gene_vega
http://localhost:9000/martservice/attributes?dataset=hsapiens_gene_vega&mart=gene_vega
http://localhost:9000/martservice/filters?dataset=hsapiens_gene_vega&mart=gene_vega
Data query
<xml> <Query processor="TSV" header="true" limit="-1" client="webbrowser"> <Dataset name="hsapiens_gene_vega" config="gene_vega_ap"> <Filter name="chromosomal_region" value="2:1000000:2000000:1,4:9000000:11000000:-1"/> <Filter name="biotype" value="protein_coding"/> <Attribute name="vega_gene_id"/> <Attribute name="vega_transcript_id"/> <Attribute name="vega_translation_id"/> <Attribute name="chromosome_name"/> <Attribute name="start_position"/> <Attribute name="end_position"/> <Attribute name="strand"/> <Attribute name="band"/> </Dataset> </Query> </xml>
Paste this piece of XML in a web browse address as:
http://localhost:9000/martservice/results?query=paste_query_xml_string_here
To read more about BioMart, refer to the recent articles describing the BioMart software and its applications.
1. Zhang J, Haider S, Baran J, Cros A, Guberman JM, Hsu J, Liang Y, Yao L, Kasprzyk A. BioMart: a data federation framework for large collaborative projects. Database (Oxford). 2011 Sep 19:2011:bar038.
2.Guberman JM, Ai J, Baran J., et al. BioMart Central Portal: An Open Database Network for the Biological Community. Database (Oxford). 2011 Sep18;2011:bar041.
3. Zhang J, Baran J, Guberman JM, Haider, S, Hsu J, Liang Y, Rivkin E, Wang J, Whitty B, Wong-Erasmus M, Yao L, Kasprzyk A. International Cancer Genome Consortium Data Portal - a One-stop Shop for Cancer Genomics Data. Database (Oxford). 2011 Sep19;2011:bar026.