Difference between revisions of "BioMart Tutorial"
m (→What is a Data Mart?) |
m (→Configuring your Mart) |
||
| Line 176: | Line 176: | ||
===Configuring your Mart=== | ===Configuring your Mart=== | ||
| − | Start MartEditor by issuing the following command under martj-0.7 folder: | + | Start MartEditor by issuing the following command under <tt>martj-0.7</tt> folder: |
cd /home/gmod/software/biomart/martj-0.7 | cd /home/gmod/software/biomart/martj-0.7 | ||
Revision as of 17:33, 29 September 2009
| {{#icon: Biomart250.png|BioMart|200|BioMart}} {{#icon: GMOD2009Europe170.png|2009 GMOD Summer School - Europe |
2009 GMOD Summer School - Europe}} | BioMart Session 2009 GMOD Summer School - Europe & Americas |
__NOTITLE__
This tutorial walks you through how to install and configure a local installation of BioMart. This tutorial was originally taught by Junjun Zhang at the 2009 GMOD Summer School - Europe & Americas. The notes and VMware image used on this page are from the Europe course.
Contents
- 1 VMware
- 2 Caveats
- 3 Introduction
- 4 System overview and installation
- 5 Build your first Mart, configure and deploy BioMart Server
- 6 Access BioMart Server via program-friendly interfaces: API and MartService
- 7 Demo: create data mart using MartBuilder
- 8 Getting support
VMware
| This tutorial was taught using a VMware system image as a starting point. If you want to start with that same system, download and install the Starting image.
See VMware for what software you need to use a VMware system image, and for directions on how to get the image setup and running on your machine. |
|
Caveats
Important Note
This tutorial describes the world as it existed on the day the tutorial was given. Please be aware that things like CPAN modules, Java libraries, and Linux packages change over time, and that the instructions in the tutorial will slowly drift over time. Newer versions of tutorials will be posted as they become available.
Introduction
BioMart is a query-oriented data management and integration system. The system uses a generic data model for data integration and storage; it can be used for any type of data and is particularly suited for complex descriptive biological data. BioMart provides several interfaces for building/executing complex queries, such as, human-friendly web-based GUI, and program-friendly API and web services.
Explore over 20 public databases through BioMart Central Portal
BioMart Central Portal (http://www.biomart.org) provides a unified interface for querying over 20 public databases with a large variety of contents.
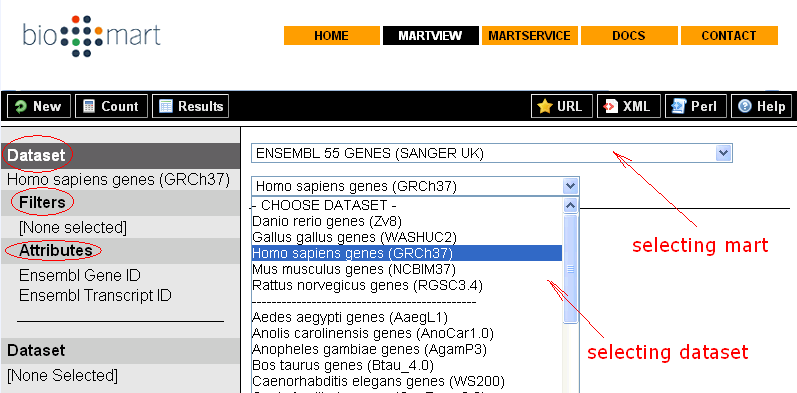
This section is intended to give you some basic ideas how BioMart helps biologists in searching data of their interests through BioMart intuitive web based GUI – MartView.
Sample queries (from http://www.biomart.org/biomart/martview):
- Retrieve Ensembl Gene ID, Chromosome Name, Gene Start (bp), Gene End (bp) of all human genes from ensembl mart (bookmark)
- Restrict the results of the previous query to region of chromosome:1, Gene Start (bp):1 and Gene End (bp):100000
- Retrieve 300bp upstream flanking sequence for Ensembl Gene: ENSG00000000419, ENSG00000000457
- How do I convert IDs? I have the following Ensembl Gene IDs from human dataset: ENSG00000000419, ENSG00000000457 and I would like HGNC symbols and RefSeq DNA IDs along with matching Affymetrix platform HG U133-PLUS-2 probes
- (Two datasets query) How do I retrieve all mouse homologues for human genes?
- (Two datasets query) Restrict the results of the previous query to human genes on chromosome 1 and mouse orthologs on chromosome 2
- (Two datasets query) Retrieve all human Ensembl Genes (output Gene ID and HGNC symbol) that are involved in a pathway with a Reactome pathway stable ID: REACT_1698 (output pathway stable ID and pathway name) (bookmark)
System overview and installation
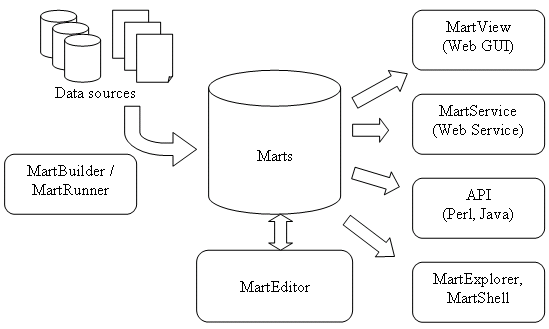
What tools are included in BioMart?
- Building Mart: MartBuilder and MartRunner
- Configuring Mart: MartEditor
- Querying Mart: Perl API, Java API, MartView (web GUI, based on Perl API), MartService (web service interface, based on Perl API), MartExplorer (based on Java API), MartShell (based on Java API)
System installation
Installing biomart-perl
Current release (0.7) of biomart-perl source code is available from CVS (password: CVSUSER):
cvs -d :pserver:cvsuser@cvs.sanger.ac.uk:/cvsroot/biomart login cvs -d :pserver:cvsuser@cvs.sanger.ac.uk:/cvsroot/biomart co -r release-0_7 biomart-perl
For this tutorial, we will use the biomart-perl source code from SVN main trunk (below).
Biomart-perl source code is available from SVN:
svn co https://code.oicr.on.ca/svn/biomart/biomart-perl/trunk biomart-perl
The svn checkout above has already been done in the VMware image at /home/gmod/software/biomart/biomart-perl.
Update your local copy of the source code:
cd /home/gmod/software/biomart/biomart-perl svn update
Prerequisites for biomart-perl
- You need to have perl version 5.6.0 or later installed first.
- biomart-perl depends on a number of perl modules, a complete list of dependencies gets listed when you run the configure script.
- You need to have apache web server and mod_perl installed.
- You will also need one database server installed. BioMart currently supports three RDBMSs: MySQL, PostgreSQL and Oracle.
Intentionally, we have left the following Perl modules for you to install:
Number::Format OLE::Storage_Lite Test::Exception Template::Plugin::Number::Format
Using apt-get:
sudo apt-get update sudo apt-get install libnumber-format-perl sudo apt-get install libole-storage-lite-perl sudo apt-get install libtest-exception-perl
Using CPAN:
sudo cpan Template::Plugin::Number::Format
Installing martj
Martj binary can be obtained as following:
cd /home/gmod/software/biomart/ wget ftp://anonymous@ftp.ebi.ac.uk/pub/software/biomart/martj_current/martj-bin.tgz tar -zxf martj-bin.tgz
After this a folder named martj-0.7 will be created under /home/gmod/software/biomart/
Prerequisites for martj
- Java 1.5 or later.
Java based tools can be launched by invoking corresponding scripts under bin directory, use *.bat for Windows, *.sh for Mac and Linux. For example, in the VMware image we can launch MartEditor as:
cd /home/gmod/software/biomart/martj-0.7 ./bin/marteditor.sh
Build your first Mart, configure and deploy BioMart Server
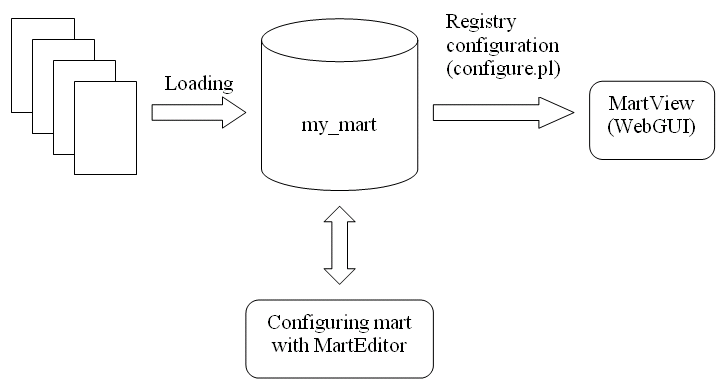
The process of deploying a BioMart Server can be logically divided into two steps: transformation and configuration. The process of transforming an existing data source into a mart database can be carried out using MartBuilder, or a user-written data convertor. The configuration; defining a view (Attributes and Filters) or multiple views on your data, is done by using MartEditor followed by a perl configure.pl script.
Workflow of creating, configuring and deploying a BioMart Server:
What is a Data Mart?
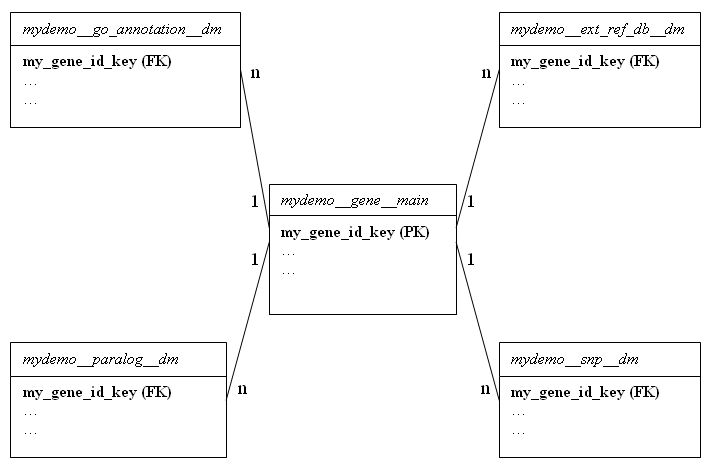
A mart is a collection of datasets. It is nearly always synonymous with a database in MySQL, or a schema in Oracle and Postgres.
A dataset is a collection of tables that follow a given naming convention. The table naming convention is dataset__content__type, where dataset is the name of the dataset, content is a free-text summary of the contents of the table, and type is either main (for main tables) or dm (for dimension tables).
Each dataset must have at least one single central table called the main table, with a type of main. This main table is involved in all queries, and will normally contain the information most frequently requested. It must have one column ending in the suffix _key which contains a unique identifier for each row, similar in function to a primary key.
A dataset may optionally have a number of dimension tables containing satellite information related to the main table. These dimension tables are recognized by having a type of dm. Each dimension table must have a column that contains values from the _key column of the main table to which the data in the dimension table is related, similar in function to a foreign key.
A dataset with a single main table and a number of dimensions looks something like this:
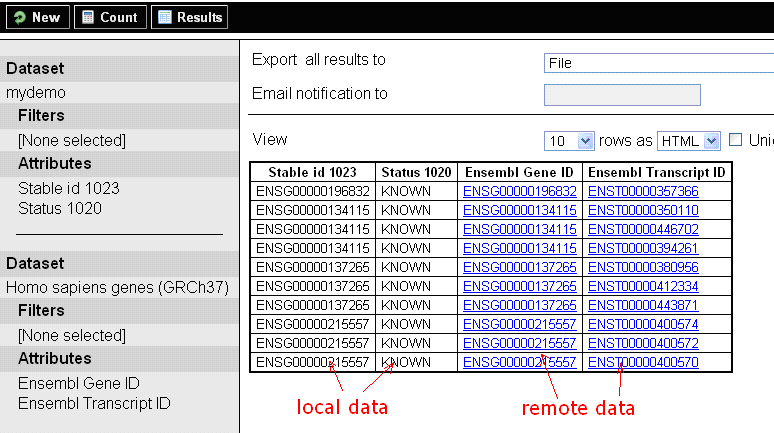
In the example above, dataset name is mydemo, it contains one main table and four dimension tables.
The set of all columns from all tables in a dataset is equivalent to the set of Attributes available on that dataset. Every Filter in a dataset is created by restricting an attribute to a particular value or range of values. Therefore filters are like the where-clause in SQL statements and attributes are like the columns listed in the select portion of a SQL statement.
One key feature of such model is its simplicity. With many fewer tables to join, the goal of high performance query is achieved. Such design is originated from the star schema in industry data warehouse. The difference is that the relation of main and dm tables is 1:n in BioMart model while it is n:1 in star schema. For that reason, the BioMart model is often referred as reversed star. What’s common is that, dimension tables (so as main table in BioMart model) are highly denormalized, i.e., related tables are merged to one table when certain rules are met. In the resulting table, values in many columns can be highly redundant. Denormalized table is also known as materialized view where join of all tables has been done and result is stored physically on the file system. Up to now, you should have realized that, the whole thing is a space-time trade-off game!
Creating your own Mart: create/load sample mart
Download demo data:
cd /home/gmod/software/biomart rm my_mart.tar.gz wget http://www.biomart.org/mart_demo.tar.gz tar -zxf mart_demo.tar.gz
Load data into mart:
cd data mysql -uroot -e 'grant all on *.* to gmod@localhost identified by "gmod"' mysql -ugmod -pgmod -e 'create database my_mart' mysql -ugmod -pgmod my_mart < my_mart.sql
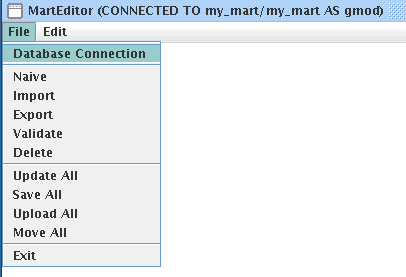
Configuring your Mart
Start MartEditor by issuing the following command under martj-0.7 folder:
cd /home/gmod/software/biomart/martj-0.7 ./bin/marteditor.sh
Please ignore if you get JDBC driver warning message.
Below lists the main menu for MartEditor:
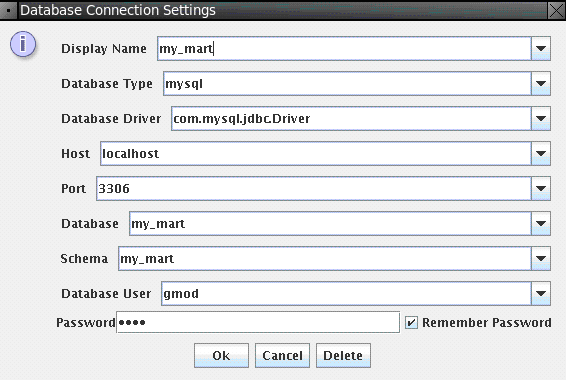
Now connect to the mart we just created, File → Database Connection, and input connection parameters as shown below:
Password is gmod.
File → Naïve, then choose dataset: mydemo
This will create a naïve configuration of the newly created dataset. For now we will just use this configuration to continue the process of setting up BioMart Web Server. Later, we will go back to MartEditor to make some adjustments and add some more stuff.
Finally, File → Export, which will save the configuration back to the meta tables in the mart we created: my_mart.
Setting the Registry
The registry file refers to the connection parameters to the data sources (ie, marts) you would like to include. This could be your own database (mart) or a publicly available mart. Several example registry files (*.xml) are available under the directory:
/home/gmod/software/biomart/biomart-perl/conf/
Here is the registry for the mart we just created: <xml>
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE MartRegistry>
<MartRegistry>
<MartDBLocation
name = "my_mart"
displayName = "My BioMart Database"
databaseType = "mysql"
host = "localhost"
port = "3306"
database = "my_mart"
schema = "my_mart"
user = "gmod"
password = "gmod"
visible = "1"
default = ""
includeDatasets = ""
martUser = ""
/>
</MartRegistry>
</xml>
We save it in my_mart.xml under biomart-perl/conf folder:
cd /home/gmod/software/biomart/biomart-perl/conf xedit my_mart.xml
Setting Web Server Configuration
biomart-perl creates a custom apache web server configuration file (httpd.conf) under biomart-perl/conf which is later used to start apache web server. What goes into this file is totally dynamic and automated. However, deployers are expected to set the path to apache binary, host name, port and apxs in biomart-perl/conf/settings.conf. The settings specified in this file are used by configure step explained in the next section.
Open conf/settings.conf with xedit and set the following:
apacheBinary=/usr/sbin/apache2 serverHost=localhost port=9002 apxs=/usr/bin/apxs2
Run Configure Script
From the biomart-perl directory, type:
cd ~/software/biomart/biomart-perl perl bin/configure.pl -r conf/my_mart.xml --clean
It will ask:
Do you want to install in API only mode [y/n] [n]:
Type n and hit Enter.
Starting and stopping Web Server
From the biomart-perl directory, to start the apache server, type:
/usr/sbin/apache2 -d $PWD -f $PWD/conf/httpd.conf
to stop the apache server, type:
kill `cat logs/httpd.pid`
Testing MartView
Now, point your web browser to:
and see if the installation went fine. Note: replace localhost with the IP address of your VM if you run web browser from your laptop's OS.
More exercises with MartEditor
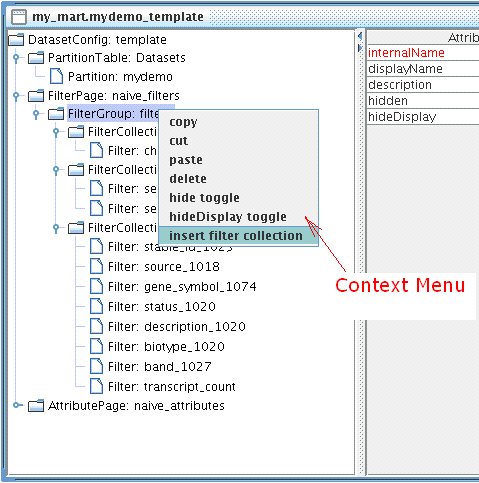
Create two new FilterCollections: Chromosome and Gene Type
Context Menu can be access by mouse right clicking any nodes in the Tree Panel. To insert a new FilterCollection, right click FilterGroup you wish to add to.
Do the following steps:
- insert a nwe FilterCollection make displayName to be Gene Type
- cut-n-paste biotype_1020 Filter to Gene Type FilterCollection
- insert a new FilterCollection, change its displayName to Chromosome
- drag-n-drop chromosome_name_1059 Filter to Chromosome FilterCollection
We can also modify some default values used in the naive configuration:
- change displayName of attribute:stable_id_1023 to Ensembl Gene ID
- set default to true for attribute:stable_id_1023
- change displayName of attribute:gene_symbol_1074 to Gene symbol
- set default to true for attribute:gene_symbol_1074
Don't forget to Export your new configure from MartEditor.
Now stop apache server, re-run configure.pl, and start apache server again. Make sure you are in /home/gmod/software/biomart/biomart-perl, then do the following:
kill `cat logs/httpd.pid` perl bin/configure.pl -r conf/my_mart.xml --clean /usr/sbin/apache2 -d $PWD -f $PWD/conf/httpd.conf
We will need to do this a few times more, so it's better to put the commands in a shell script:
cd /home/gmod/software/biomart/biomart-perl xedit restart.sh
Copy and paste, then save.
Make it executable by everyone:
chmod +x restart.sh
Next time we need to reconfig the server, we do:
cd /home/gmod/software/biomart/biomart-perl ./restart.sh
Go to http://localhost:9002/biomart/martview to check out the new FilterCollections we just created.
Make a dropdown list for Chromosome name Filter
Right-click Chromosome name Filter, from the Context Menu choose make drop down, you are done!
If you want to allow multiple options to be selected in this drop down list, simply set multipleValues to 1, export configuration, and reconfigure MartView.
Export new configure.
Now stop apache server, re-run configure.pl, and start apache server again.
cd /home/gmod/software/biomart/biomart-perl/ ./restart.sh
Go to http://localhost:9002/biomart/martview to check out the change for Chromosome name filter.
Configure links between datasets (ie, federation)
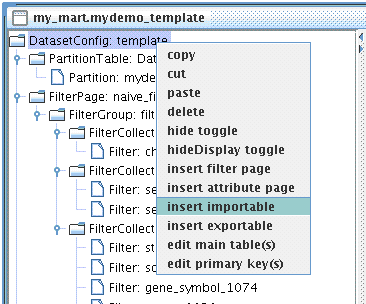
In BioMart, a link is built through a pair of Exportable and Importable, each defined in one of the two to-be-linked datasets.
We can think of an Exportable is an Attribute (or an Attribute list) which one dataset exports to the other dataset to fetch related data records. Similarly, an Importable can be seen as a Filter, one dataset takes Exportable from the other dataset and apply it to its own Filter.
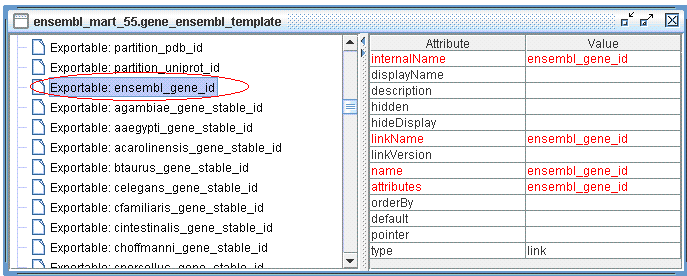
Let's look at an example: the mydemo dataset can be linked with hsapiens_gene_ensembl in Enseml Gene mart by the common Ensembl Gene ID field. We can define an Exportable in hsapiens_gene_ensembl, and an Importable in mydemo.
hsapiens_gene_ensembl already has an Exportable defined, see below:
Useful tip: you can always connect to Ensembl Mart with MartEditor to learn how Filters and Attributes are defined.
| Ensembl Mart MySQL connection parameters | |
|---|---|
| Host | martdb.ensembl.org |
| Port | 5316 |
| User | anonymous |
| Databases | ensembl_mart_55 |
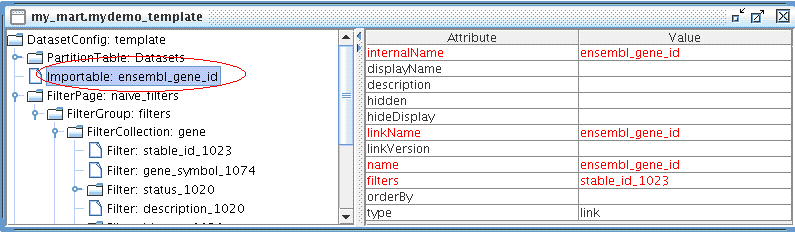
Now let's create an Importable for mydemo dataset.
The Importable should look like this:
Don't forget to Export your configuration to mart: File → Export
Now, we have to add hsapiens_gene_ensembl dataset in the registry, together with mydemo.
Here is what the new registry looks like: <xml>
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE MartRegistry>
<MartRegistry>
<MartDBLocation
name = "my_mart"
displayName = "My BioMart Database"
databaseType = "mysql"
host = "localhost"
port = "3306"
database = "my_mart"
schema = "my_mart"
user = "gmod"
password = "gmod"
visible = "1"
default = ""
includeDatasets = ""
martUser = ""
/>
<MartDBLocation
name = "ensembl_gene"
displayName = "Ensembl Gene"
databaseType = "mysql"
host = "martdb.ensembl.org"
port = "5316"
database = "ensembl_mart_55"
schema = "ensembl_mart_55"
user = "anonymous"
password = ""
visible = "1"
default = ""
includeDatasets = "hsapiens_gene_ensembl"
martUser = ""
/>
</MartRegistry>
</xml> Now stop apache server, re-run configure.pl, and start apache server again.
cd /home/gmod/software/biomart/biomart-perl/ ./restart.sh
Finally, you can test queries against federated datasets at http://localhost:9002/biomart/martview.
Access BioMart Server via program-friendly interfaces: API and MartService
Perl API
After set a query in MartView, you can click the Perl button (top right corner), you will get a piece of automatically generated Perl code. With few simple modifications, you can run the code to query dataset through Perl API. Here is a sample query
Let's copy and paste the perl code in xedit, save the code under /home/gmod/software/biomart/biomart-perl/scripts.
cd /home/gmod/software/biomart/biomart-perl/scripts xedit myApiTest.pl
Add this line to include Perl libraries at the top of the code: <perl>use lib '/home/gmod/software/biomart/biomart-perl/lib';</perl>
Modify this line to set the correct registry file: <perl>my $confFile = '/home/gmod/software/biomart/biomart-perl/conf/my_mart.xml';</perl>
Run it as:
perl myApiTest.pl
MartService
MartService provides a program-friendly interface for end-users and third-party tools to interact with a BioMart Server. There are a few systems (eg. Taverna, Galaxy and biomaRt R package) have implemented plugins based on MartService.
Get Results
The following request is used to retrieve data from a BioMart database. An XML based query containing attributes, filters and datasets is POSTED to a target BioMart web server which returns either results or number of entries based on the request.
localhost:9002/biomart/martservice?query=<QUERY_XML>
A Query XML example: <xml>
<?xml version="1.0" encoding="UTF-8"?> <!DOCTYPE Query> <Query virtualSchemaName = "default" formatter = "TSV" header = "0" uniqueRows = "0" count = "" datasetConfigVersion = "0.6" > <Dataset name = "mydemo" interface = "default" > <Filter name = "chromosome_name_1059" value = "1"/> <Attribute name = "stable_id_1023" /> <Attribute name = "gene_symbol_1074" /> <Attribute name = "chromosome_name_1059" /> <Attribute name = "seq_region_start_1020" /> <Attribute name = "seq_region_end_1020" /> <Attribute name = "source_1018" /> </Dataset> </Query>
</xml>
Useful tip:
- To retrieve an XML Query from any BioMart Web interface (MartView), hit the XML button after making your selection of database, datasets, attributes and filters
Save the above query XML in query.xml and put it under /home/gmod/software/biomart/biomart-perl/scripts.
cd /home/gmod/software/biomart/biomart-perl/scripts xedit query.xml
Edit webExample.pl so that path points to your own server:
my $path="http://localhost:9002/biomart/martservice?";
Now run:
perl webExample.pl query.xml
Get Metadata
The requests described in this section are used to retrieve which marts, datasets, attributes, filters and formatters are available on a particular BioMart web server.
Demo: create data mart using MartBuilder
Prepare source data
cd /home/gmod/software/biomart/data mysql -ugmod -pgmod -e 'create database student' mysql -ugmod -pgmod student < student.sql
Create student_mart
We now start MartBuilder:
cd /home/gmod/software/biomart/martj-0.7 ./bin/martbuilder.sh
First add the source schema, Schema → Add
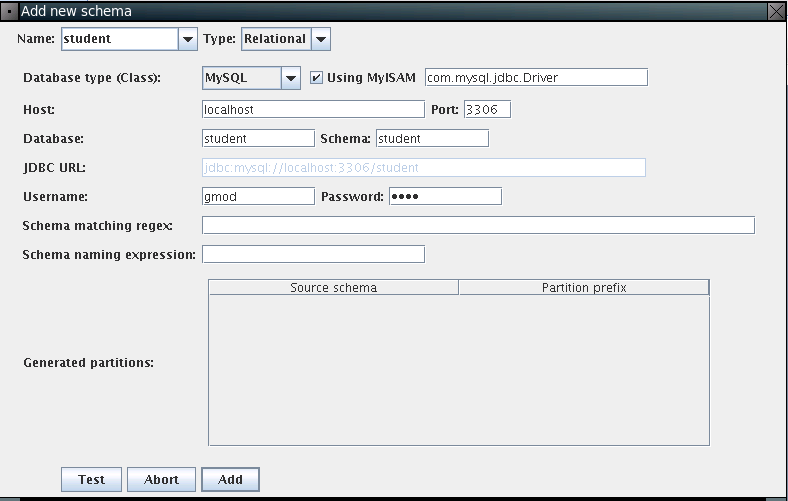
Here, please input connection parameters:
Now you should be able to see the student schema:
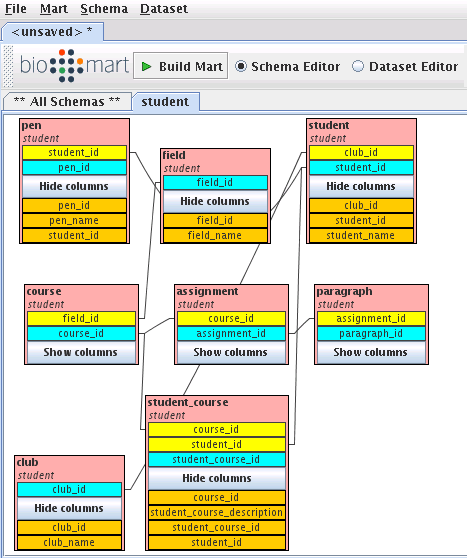
Right-click on student table, then choose create dataset for student:
We are now going to transform the source data into target dataset, but before that, we have to create a target database:
mysql -ugmod -pgmod -e 'create database student_mart'
Also we have to have MartRunner running. Let's run it over port 8888:
cd /home/gmod/software/biomart/martj-0.7 ./bin/martrunner.sh 8888
MartBuilder will send the transformation SQL to MartRunner through port 8888, and MartRunner will execute the transformation SQL. Usually, MartBuilder and MartRunner run on different machines.
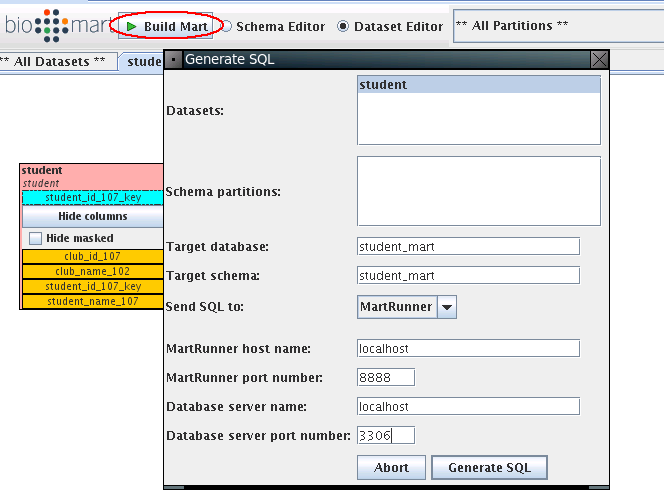
We go back to MartBuilder, clike Build Mart:
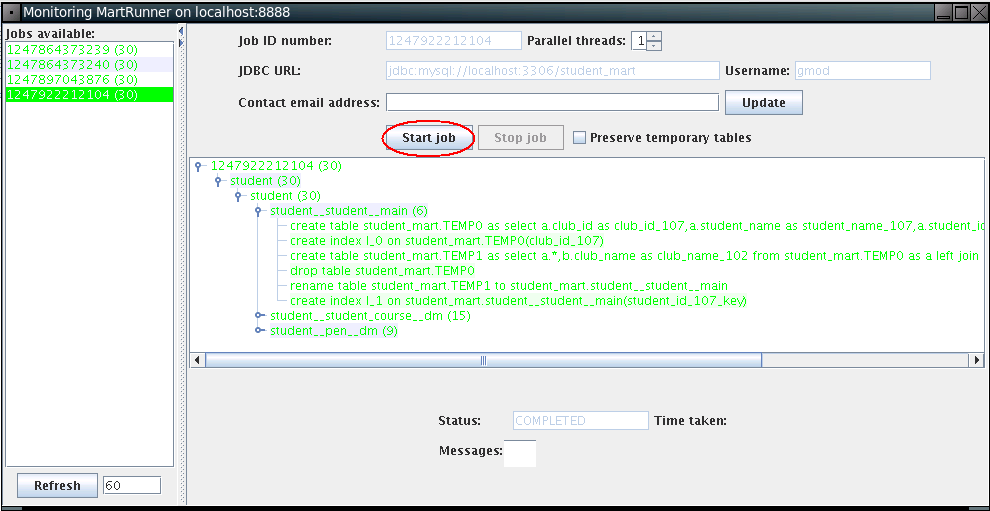
The MartRunner monitor window will show up as below. Click Start job to build student_mart.
Configure and deploy student_mart
- Start MartEditor; connect to student_mart; Naive; Export
- Add one more MartDBLocation entry in my_mart.xml (under /home/gmod/software/biomart/biomart-perl/conf/) pointing to student_mart database
<xml>
<MartDBLocation
name = "student_mart"
displayName = "My Student Database"
databaseType = "mysql"
host = "localhost"
port = "3306"
database = "student_mart"
schema = "student_mart"
user = "gmod"
password = "gmod"
visible = "1"
default = ""
includeDatasets = ""
martUser = ""
/>
</xml>
- Restart your BioMart Server:
cd ~/software/biomart/biomart-perl/ ./restart.sh
- Query student_mart using MartView
Getting support
- Further documentation from http://www.biomart.org
- Mailing list mart-dev@ebi.ac.uk