Difference between revisions of "JBrowse2 Tutorial PAG 2023"
(→Adding a gene track from tabix-indexed GFF) |
(→Adding a gene track from a JBrowse (NCList) track) |
||
| Line 114: | Line 114: | ||
==Adding a gene track from a JBrowse (NCList) track== | ==Adding a gene track from a JBrowse (NCList) track== | ||
| + | |||
| + | Another sort of data that can be used with JBrowse 2 is data that is currently being used with JBrowse 1. At WormBase there is a JBrowse 1 instance with many tracks. We'll use one of those as an example here to load a track that has just protein coding genes: | ||
<span class="enter"> | <span class="enter"> | ||
| Line 122: | Line 124: | ||
Note that the --load option isn't required for URLs. | Note that the --load option isn't required for URLs. | ||
| − | |||
| − | |||
| − | |||
| − | |||
[[File:protein_coding_genes.png|800px]] | [[File:protein_coding_genes.png|800px]] | ||
Revision as of 18:46, 2 December 2022
Contents
- 1 Prerequisites
- 2 Initializing JBrowse
- 3 Adding a reference sequence
- 4 Adding a gene track from tabix-indexed GFF
- 5 Adding a gene track from a JBrowse (NCList) track
- 6 Adding variant data from a tabix-indexed VCF
- 7 Adding quantitative data from a BigWig
- 8 Using JEXL to modify the display
- 9 Synteny
- 10 Adding text search indexes
- 11 Using the admin-server
Prerequisites
- NodeJS
Installed using the instructions on Nodejs.org:
curl -fsSL https://deb.nodesource.com/setup_18.x | sudo -E bash - &&sudo apt-get install -y nodejs
- A web server (Apache2 in this instance, but any will do). I enabled the "userdir" mod so we could all use the same machine for the tutorial:
sudo a2enmod userdir sudo /etc/init.d/apache2 restart
Things done just for this tutorial
- A script to create several users with
public_htmldirectories (link for when it exists) - Already installed the JBrowse command line interface (CLI) via the directions (i.e.,
sudo npm install -g @jbrowse/cli) - Installed bgzip, tabix, samtools and minimap2 via apt:
sudo apt-get install samtools tabix minimap2. - Created a bgzipped and samtools faidx'ed FASTAs file for C. elegans and C. brenneri.
- Created a "Genes only" C. elegans GFF file (
gzip -dc c_elegans.PRJNA13758.WS286.annotations.gff3.gz | grep "\tWormBase\t" > c_elegans.genes.gff3
Initializing JBrowse
First, use ssh to connect to the instance we have set up for this tutorial, tutorialpag30.jbrowse.org. Do this with the user name and password you got from one of us (we have 50 users configured--hopefully that will be enough!):
ssh username@tutorialpag30.jbrowse.org
and supply the password. When you log in, you'll be in your user's home directory, where there is nothing but a public_html directory. We'll use the JBrowse CLI to initialize a new JBrowse instance:
jbrowse create public_html --force
Note that the "--force" is necessary here because the public_html directory isn't empty and the create script doesn't want to accidentally delete any existing files. We're safe though. Now change to that directory, cd public_html and do a file list to make sure it looks right:
This is all of the software required to run JBrowse, plus soft links to files we are going to use in the tutorial. If we now navigate to the tutorial machine's website with the username on the slip provided at the beginning, you should see a page indicating that JBrowse was installed but not configured: http://tutorialpag30.jbrowse.org/~userXX. (Of course, substitute in your username in the URL)
To make sure it really works, we can click on the Volvox (not really Volvox) data set.
Adding a reference sequence
The first thing we need to do is add a reference sequence. There is a samtools/faidx indexed fasta file already in your public_html directory. To create this indexed reference sequence, the fasta was downloaded from the WormBase ftp site, and after uncompressing it, it was bgzipped and then indexed with SAMTools:
bgzip c_elegans.PRJNA13758.WS286.genomic.fa samtools faidx c_elegans.PRJNA13758.WS286.genomic.fa.gz
To tell JBrowse about the new assembly, we can use the jbrowse CLI:
jbrowse add-assembly c_elegans.PRJNA13758.WS286.genomic.fa.gz \
--displayName "C. elegans N2" \
--name c_elegans_PRJNA13758 \
--type bgzipFasta \
--load inPlace \
--refNameAliases test_data/ce_aliases.txt
The command will probably generate some warnings about the locations of the files, but the apache server is configured to use them where they are (ie FollowSymLinks was added to the configuration).
In this command, --displayName is what will appear in the user interface, --name is what will be used in future configuration options to refer to this assembly, --type refers to how the fasta file was indexed, and --load inPlace tells the CLI to leave the files where they are (other options include "copy", "symlink" and "move"). The --refNameAliases option gives options for how chromosomes are named; that will be covered in more detail below. Note that we don't specify the location of the .fai and .gzi files in this command; the CLI will assume the names from the name of the compressed fasta file (it is possible to specify them on the command line too if they aren't "guessable"). Copy this command and run it in the public_html directory. After doing that, go to the web browser and if you aren't on the splash screen, select "Return to splash screen" from the file menu, and start a new session and then launch a LinearGenomeView. You should get a dialog to open an assembly with "C. elegans N2" as the only option.
Go ahead and open chromosome I.
Adding a gene track from tabix-indexed GFF
GFF3 is a very common file format for defining genome features. The GFF3 file we're using today is based on the on created by WormBase for every release. Here is a sample:
I WormBase gene 4116 10230 . - . ID=Gene:WBGene00022277;Name=WBGene00022277;locus=homt-1;sequence_name=Y74C9A.3;biotype=protein_coding;so_term_name=protein_coding_gene;curie=WB:WBGene00022277;Alias=homt-1,Y74C9A.3 I WormBase mRNA 4116 10230 . - . ID=Transcript:Y74C9A.3.1;Parent=Gene:WBGene00022277;Name=Y74C9A.3.1;wormpep=CE28146;locus=homt-1;uniprot_id=Q9N4D9 I WormBase three_prime_UTR 4116 4220 . - . Parent=Transcript:Y74C9A.3.1 I WormBase CDS 4221 4358 . - 0 ID=CDS:Y74C9A.3;Parent=Transcript:Y74C9A.3.1;Name=Y74C9A.3;prediction_status=Confirmed;wormpep=CE28146;protein_id=CCD68263.1;locus=homt-1;uniprot_id=Q9N4D9 I WormBase intron 4359 5194 . - . Parent=Transcript:Y74C9A.3.1;Note=Confirmed_EST yk1692c07.3 %3B Confirmed_EST OSTR037H1_1 %3B Confirmed_EST elegans_PE_SS_GG2157%7Cc1_g1_i1 %3B Confirmed_EST elegans_PE_SS_GG2157%7Cc1_g1_i1 %3B Confirmed_EST adult_Nanopore_Roach_35350 %3B Confirmed_EST adult_Nanopore_Roach_35350 %3B Confirmed_EST adult_Nanopore_Roach_35350 %3B Confirmed_EST adult_Nanopore_Roach_35350 %3B I WormBase CDS 5195 5296 . - 0 ID=CDS:Y74C9A.3;Parent=Transcript:Y74C9A.3.1;Name=Y74C9A.3;prediction_status=Confirmed;wormpep=CE28146;protein_id=CCD68263.1;locus=homt-1;uniprot_id=Q9N4D9
One of the easiest ways to use GFF3 with JBrowse is to use a tabix indexed bgzipped file. Generally, before creating the tabix index, GFF3 files have to be sorted first by the reference sequence (ie, the chromosome name, in column 1) and then by the starting coordinate (colunn 4). Here is a magic incantation for doing that on the Linux command line (sort and then pipe the result to bgzip):
sort -t"`printf '\t'`" -k1,1 -k4,4n c_elegans.genes.gff3 |bgzip > c_elegans.genes.sorted.gff3.gz
and then tabix indexing it:
tabix c_elegans.genes.sorted.gff3.gz
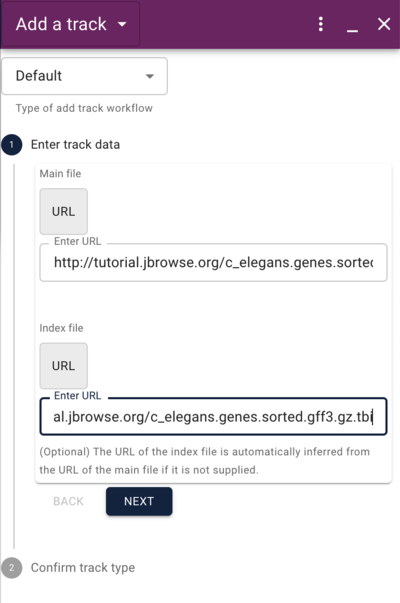
To save time, we placed both of these file in your public_html directory as well. To use the CLI to add a GFF track, do this:
jbrowse add-track c_elegans.genes.sorted.gff3.gz \
--name Genes \
--description "Curated genes from WormBase" \
--load inPlace
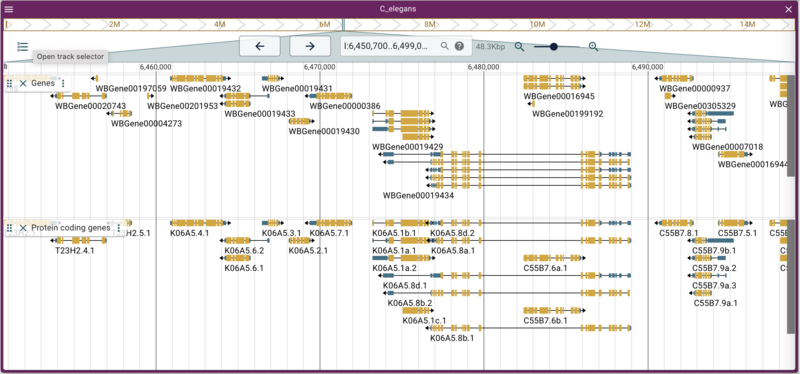
Where the options are the same as before, with the --description option to provide information about what the track is. Reload the page in the web browser and open the track selector if it isn't already open, and select the "Genes" track. Note that when you hover your mouse over the Genes track checkbox, the description appears in a tooltip.
Adding a gene track from a JBrowse (NCList) track
Another sort of data that can be used with JBrowse 2 is data that is currently being used with JBrowse 1. At WormBase there is a JBrowse 1 instance with many tracks. We'll use one of those as an example here to load a track that has just protein coding genes:
jbrowse add-track https://s3.amazonaws.com/agrjbrowse/MOD-jbrowses/WormBase/WS286/c_elegans_PRJNA13758/tracks/Curated%20Genes%20\(protein%20coding\)/{refseq}/trackData.jsonz \ --name "Protein coding genes" \ --description "Only protein coding genes from WormBase"
Note that the --load option isn't required for URLs.
Side note: finding JBrowse 1 data
CORS
Difference between web and desktop
Adding variant data from a tabix-indexed VCF
jbrowse add-track https://storage.googleapis.com/elegansvariation.org/releases/current/WI.current.soft-filtered.vcf.gz \ --name Variants
https://storage.googleapis.com/elegansvariation.org/releases/current/WI.current.soft-filtered.vcf.gz
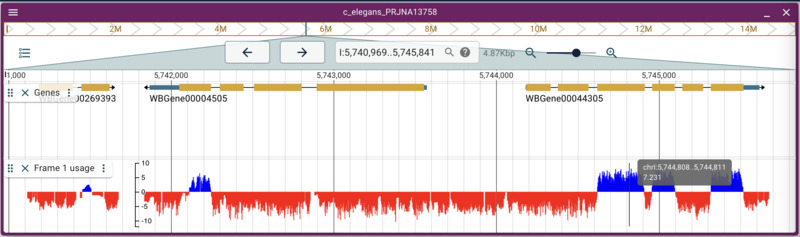
Adding quantitative data from a BigWig
jbrowse add-track https://data.broadinstitute.org/compbio1/PhyloCSFtracks/ce11/latest/PhyloCSF+1.bw \ --name "Frame 1 usage"
Aliases!
https://data.broadinstitute.org/compbio1/PhyloCSFtracks/ce11/latest/PhyloCSF+1.bw
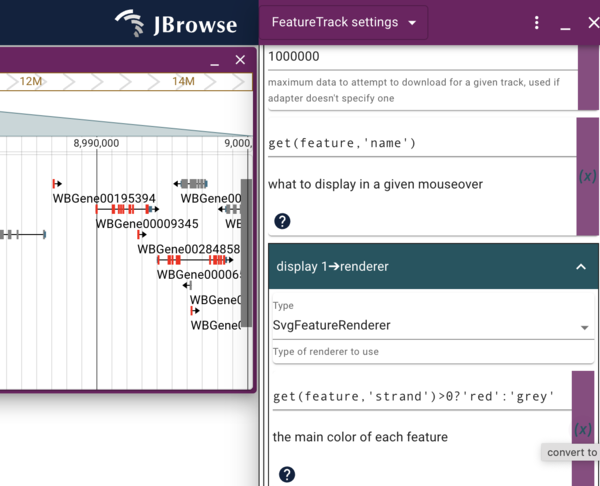
Using JEXL to modify the display
Copy a track so we can make local changes to the settings.
Dynamically changing the color
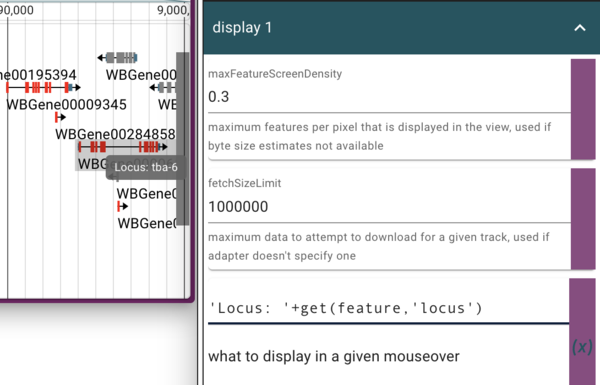
Dynamically changing the mouseover text
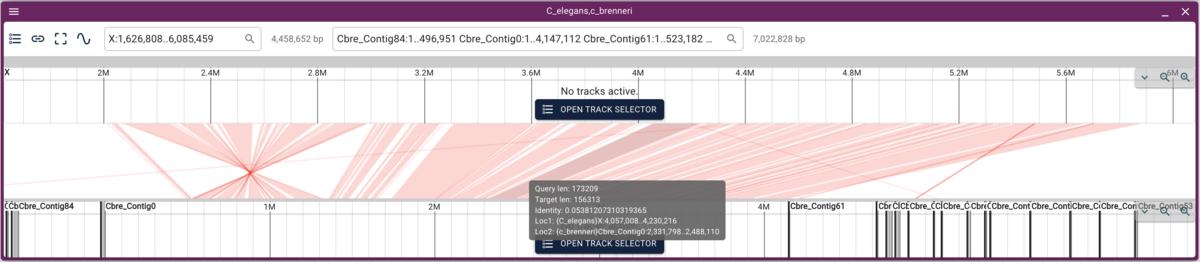
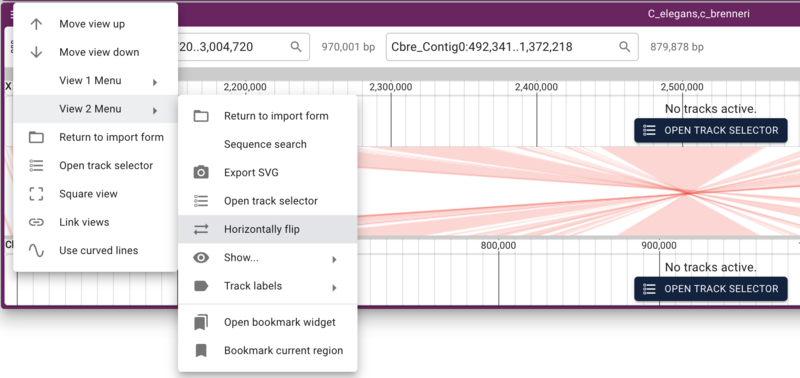
Synteny
Getting the data
To compare two genomes, first we need a second genome. Fortunately, WormBase.org provides several assemblies for species related to C. elegans. For this tutorial, we'll use C. brenneri. As before, we create a new assembly in JBrowse with the indexed fasta files provided on the tutorial machine:
jbrowse add-assembly c_brenneri.PRJNA20035.WS287.genomic.fa.gz \
--displayName "C. brenneri" \
--name c_brenneri_PRJNA20035 \
--type bgzipFasta \
--load inPlace
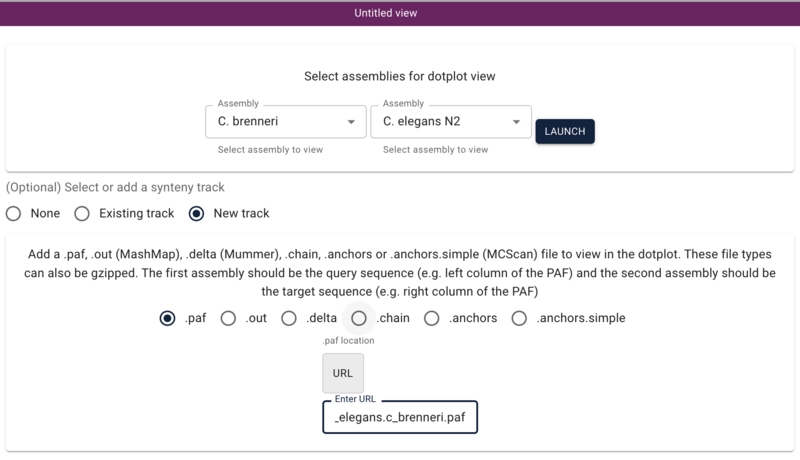
Next we need some analysis result that compares the two genomes. The tool minimap2 is a fairly lightweight application that will do a fast comparison and generate a PAF file (other formats that JBrowse supports include anchors files from MScanX, out files from MashMap, and delta files from Mummer). We can all do this now, and hopefully not crash the machine we're running on:
minimap2 c_elegans.PRJNA13758.WS286.genomic.fa.gz c_brenneri.PRJNA20035.WS287.genomic.fa.gz > c_elegans.c_brenneri.paf
Configuring with jbrowse admin-server
jbrowse add-track c_elegans.c_brenneri.paf \
--assemblyNames c_brenneri_PRJNA20035,c_elegans_PRJNA13758 \
--description "A minimap2 comparison of C. elegans and C. brenneri" \
--load inPlace \
--name "C. elegans/C. brenneri Synteny"
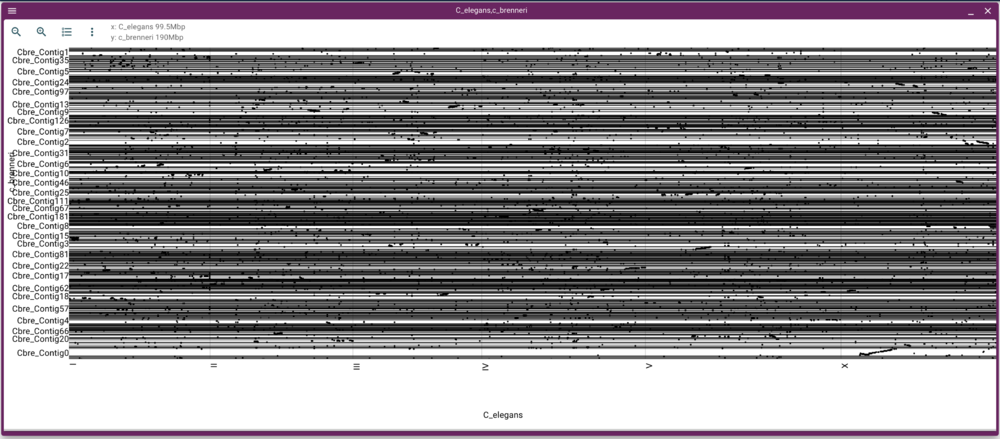
Using dotplot and synteny views
Adding text search indexes
Creating trix index
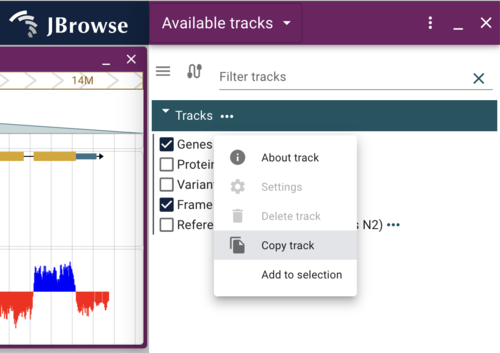
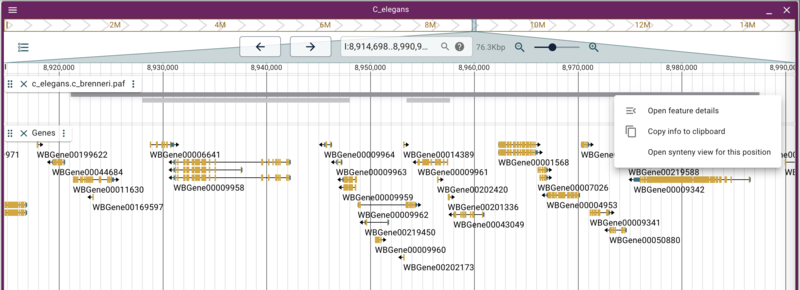
The jbrowse CLI provide a tools to create text indexes of many of the data sources we used, like tabix indexed files. Note that it does not index JBrowse 1 (NClist) data; that we'll do below. We can create a searchable index for the Genes track we created but we should exclude the VCF because it's very big and indexing it wouldn't help our users. To do that, we first have to find the trackId of the Genes track. Click on the ... after the name of the Genes track and select "About track", and copy the value of the config.trackId. It will look something like c_elegans.genes.sorted.gff3. Now on the command line in the public_html directory, run the command
jbrowse text-index --attributes=Name,ID,locus --tracks=<genes trackId>
WARNING: if you don't exclude the VCF file, it will take a very long time to run, as it will fetch the large VCF file from Google and index it.
The default value for the attributes flag are "Name,ID" so it will index the GFF attributes that have those tags. For this command, we add the "locus" attribute, because WormBase puts the "human readable" name in that attribute.
Adding a JBrowse 1 name index
Using the admin-server
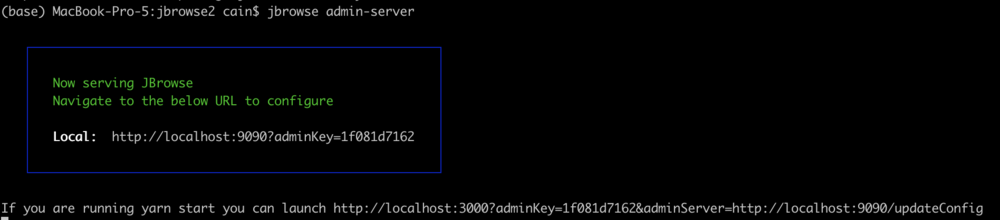
We can also run the JBrowse admin-server, which looks just like JBrowse proper, but has an extra admin menu. Important note: The admin server is NOT meant to be left running; it is not particularly secure, so if you leave it up, somebody might start messing with your site. To start the admin server, we change to the directory where JBrowse will be served from (public_html) and run the jbrowse command to start it:
jbrowse admin-server -p YYYY
where YYYY is the port number on the username/password card we passed out at the beginning. When we execute that command, we get a message in the terminal that it started up and gives us some URLs to use to access the server. It will look something like this:
The part we need is the adminKey. In a browser window, enter a URL that looks like this: http://tutorialpag30.jbrowse.org:YYYY?adminKey=yourkey
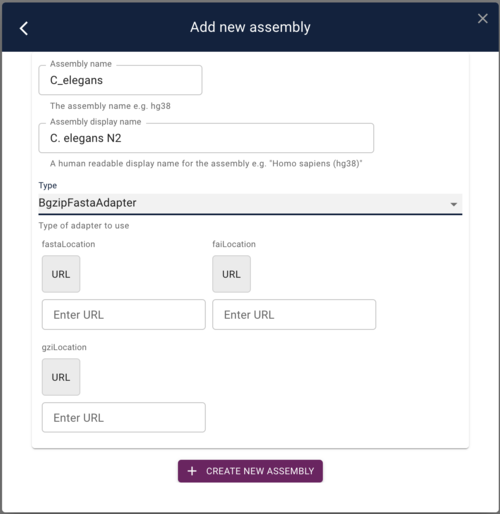
Using the admin-server to add an assembly
The first thing we need to do is add a reference sequence. There is already one prepared and on the web server for C. elegans and it is at
http://tutorialpag30.jbrowse.org/c_elegans.PRJNA13758.WS286.genomic.fa.gz http://tutorialpag30.jbrowse.org/c_elegans.PRJNA13758.WS286.genomic.fa.gz.fai http://tutorialpag30.jbrowse.org/c_elegans.PRJNA13758.WS286.genomic.fa.gz.gzi
To add this as a reference sequence to JBrowse, click on the "Start a new session" and then on the resulting page, select "Open assembly manager" from the Admin menu. In the dialog that opens, click the "Add new assembly" button. Finally, in add assembly dialog, put something useful in the "Assembly Name" field and then select "BgzipFastaAdapter" from the "Type" menu. At that point, the dialog will change slightly to give you places to put in the above three URLs:
Copy and paste those URLs in to the appropriate fields and then click "Save new assembly."
Note: this is one place where the web version of JBrowse with the admin server is slightly different from the Desktop version: if we were using the desktop version, the above dialog would have also given the option for finding the files on a local hard drive rather than only allowing URLs. Another note: In order for the above URLs to work with a web instance of JBrowse that isn't on the "same" server (where different ports == a different server), CORS (cross origin resource sharing) had to be enabled for the web server (in this case apache). If you want to do the same thing for a server you control, google "enable CORS <your server software name>" to find directions.
Using the admin-server to add a GFF track
http://tutorialpag30.jbrowse.org/c_elegans.genes.sorted.gff3.gz http://tutorialpag30.jbrowse.org/c_elegans.genes.sorted.gff3.gz.tbi
Using the admin-server to add a synteny track
http://tutorialpag30.jbrowse.org/c_elegans.c_brenneri.paf