CMap
From GMOD
Contents
Description
CMap is a web-based tool that allows users to view comparisons of genetic and physical maps. The package also includes tools for curating map data.
Requirements
- Perl 5.6.0 or higher
- A web server with CGI capabilites (e.g., Apache 1.x or 2.x)
- A relational database, e.g. MySQL (4.0+), Sybase Adaptive Server Enterprise, PostgreSQL, SQLite or Oracle (9.x)
- libgd 2.0.28 and GD.pm 2.15
- Various Perl modules available on CPAN
Documentation
- README
- Changes
- INSTALL
- ADMINISTRATION
- CODE_OVERVIEW
- TODO
- Attributes and Cross-references
- Description
- User Tutorial
Downloads
Demo & Screenshots
See the Gramene Project for a production version.
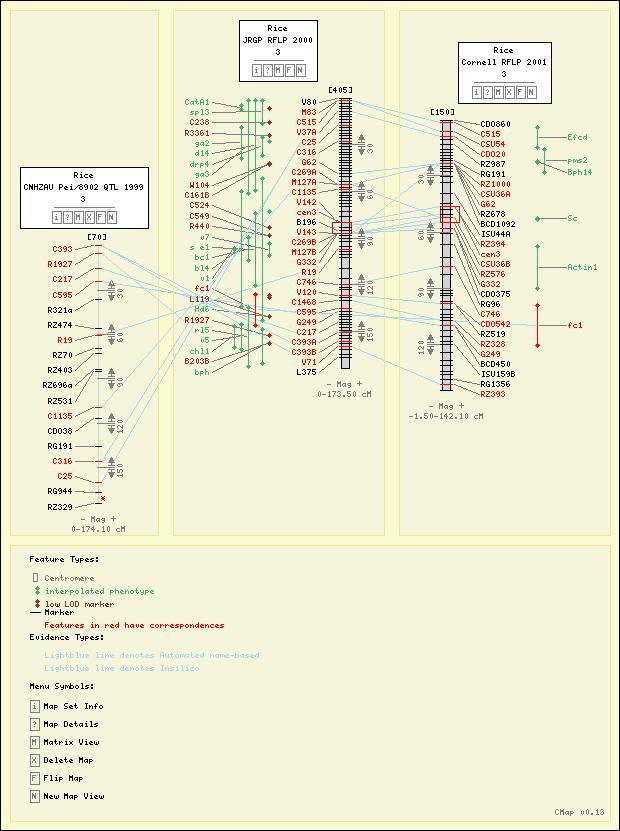
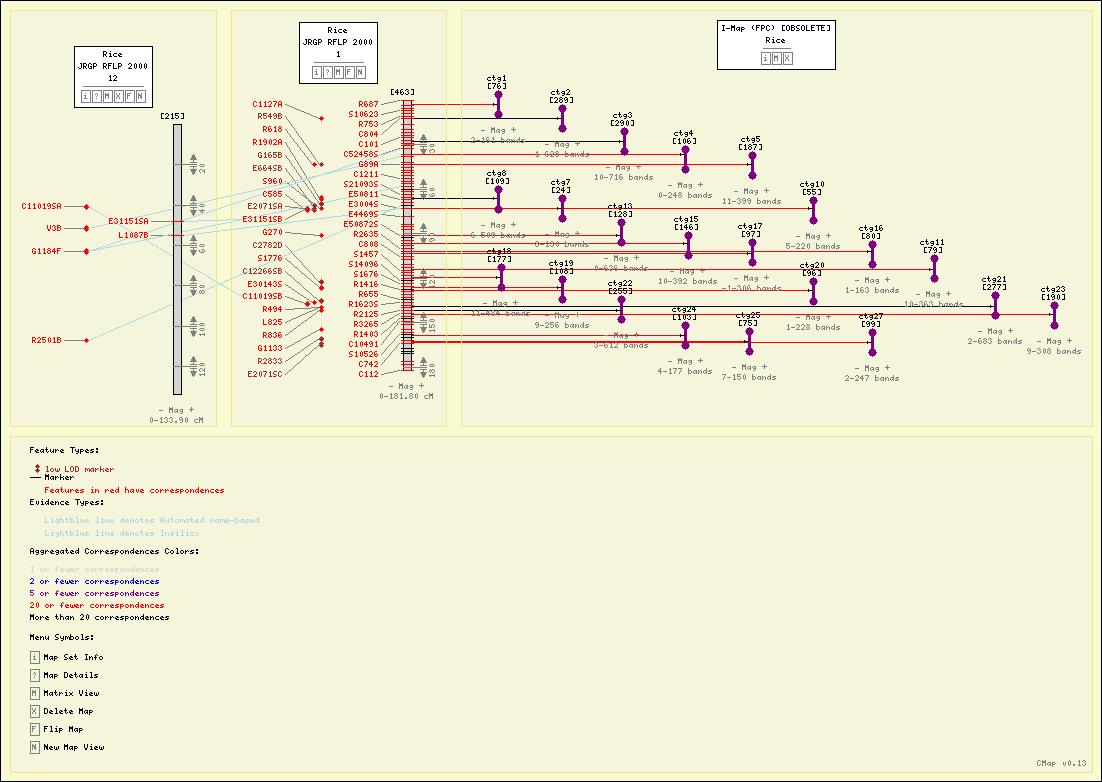
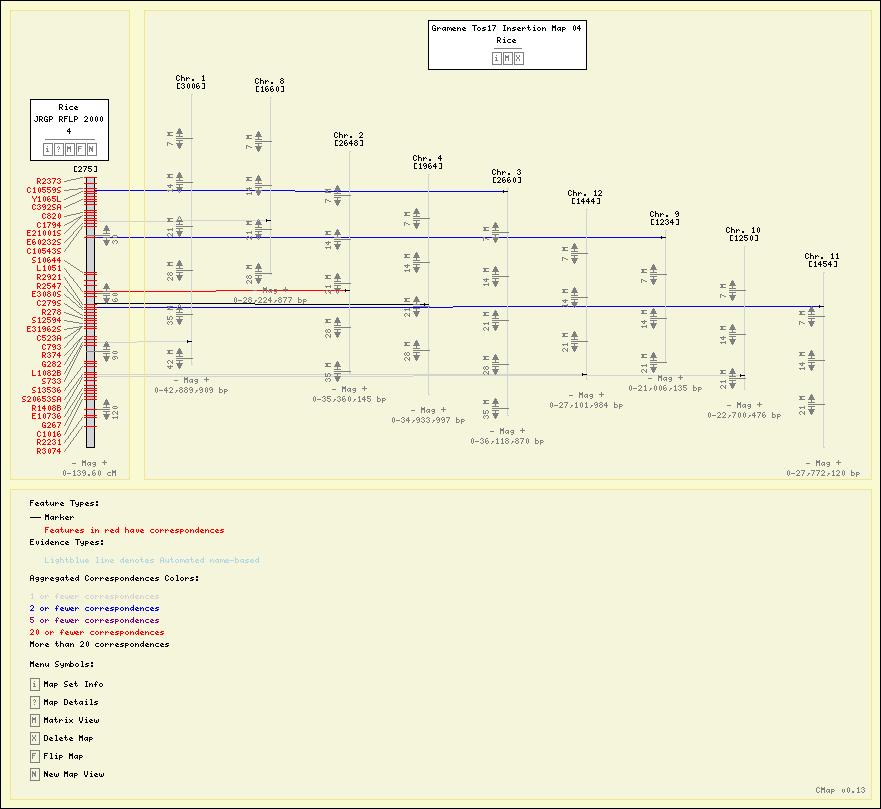
Screenshots
Here are some sample maps from the Gramene Project.
Sites Using CMap
- Gramene: Comparative mapping resource for grasses (plant)
- NCGR's Legume Information System: A publicly accessible legume resource that will integrate genetic and molecular data from multiple legume species and enable cross-species comparisons
- BeeBase: The honey bee genome database
- BASC: Brassica database
Code
Patches
- See the SourceForge Patch List.
Mailing List
- gmod-cmap@lists.sourceforge.net: Discussion of development, installation problems, etc.
- gmod-cmap-commits@lists.sourceforge.net: Notification of CVS activity
- Subscribe
- Archives