GBrowse syn
__NOTITLE__
The Generic Synteny Browser
GBrowse_syn, or the Generic Synteny Browser, is a GBrowse-based synteny browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later). Working examples can be seen at TAIR, WormBase, and SGN.
- Mature release
- Active development
- Active support
Contents
Notes on Whole Genome Alignment Data
The focus of this documentation is on the GBrowse_syn application. However, how to generate whole genome alignments, identify orthologous regions, etc, are the subject of considerable interest, so some background reading is listed below:
- Primer on Hierarchical Genome Alignment Strategies
- article on PECAN and ENREDO
- all about PECAN
- Information about EnsEMBL's compara pipeline
Installation
- GBrowse_syn uses much of the same infrastructure as GBrowse, in terms of species databases, configuration files, perl libraries, etc.
- It comes as part of the GBrowse distribution, version 1.69 and later (GBrowse installation info...).
- It differs in that databases and configuration for individual species are linked together via a central configuration file and a joining database that contains reciprocal alignments between all species represented in the browser.
- We recommend using the most up-to-date version of the application (v. 1.7 or later) either with the GBrowse netinstall script (use the -d option) or with a source code installation from SVN.
Configuration
- Configuration of GBrowse_syn is much the same as for GBrowse, with database and display options controlled by a configuration file
- GBrowse_syn uses a main configuration file for general options plus an individual configuration for each species represented in the mutliple sequence alignments
- Details....
The Alignment data
- GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments
- There is an additional GBrowse database for each species represented in the alignments
- The species' databases are configured in the same way as a regular GBrowse installations
- Details...
User interface
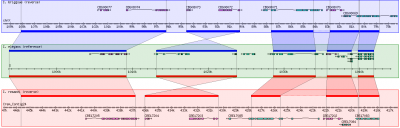
- The overall look of Gbrowse_syn resembles GBrowse but has some key differences to accomodate the more complex comparative genome data.
- GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species
- There is no upward limit of the number of species that can be displayed.
- Details...
Support
| Mailing List Link | Description | Archive(s) | |
|---|---|---|---|
| GBrowse & GBrowse_syn | gmod-gbrowse | GBrowse and GBrowse_syn users and developers. | Gmane, Nabble (2010/05+), Sourceforge |
| gmod-gbrowse-cmts | Code updates. | Sourceforge |
Presentations and Workshops
- Challenges in Comparative Genome Browsing - Presented by Sheldon McKay at the European Bioinformatics Institute, Hinxton, UK.
- Comparative Genomics with GBrowse_syn - Presentation by Sheldon McKay at the SMBE 2009 GMOD Workshop on using GBrowse_syn for comparative genomics.
- GBrowse_syn at PAG - Presentation by Sheldon McKay at the Plant and Animal Genomes meeting, San Diego, CA, USA.
- November 2007 - Sheldon McKay's presentation on GBrowse_syn at the November 2007 GMOD Meeting.
Logo
The GBrowse_syn logo was created by Darek Lakey, a participant in the Spring 2010 Logo Program, while a design student at Linn-Benton Community College.