Difference between revisions of "GBrowse syn"
m (Fixing URLs) |
m (Rearranging page) |
||
| Line 1: | Line 1: | ||
__NOTITLE__ | __NOTITLE__ | ||
| − | + | [[File:GBrowse_syn_logo.png|center|400px|alt=GBrowse_syn logo]] | |
| − | | | + | |
| − | | | + | |
| − | + | ||
| − | + | ||
| − | + | <div class="componentBox"> | |
| − | + | <div class="compBoxHdr">Status</div> | |
| − | + | *Mature release | |
| + | *maintenance | ||
| + | *active support | ||
| + | <div class="compBoxHdr">Resources</div> | ||
| + | *[http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ Download] | ||
| + | *{{2008SurveyLink|#GBrowse_syn|2008 Survey}} | ||
| + | {{ComponentBoxSectionHeader | Events}} | ||
| + | {{SchoolBoxItem|GBrowse_syn}} | ||
| + | </div> | ||
| − | + | ==About Generic Synteny Browser (GBrowse_syn)== | |
| − | + | GBrowse_syn, or the Generic Synteny Browser, is a [[GBrowse]]-based [[synteny]] browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later). | |
| − | + | ||
| − | + | ===Screenshots=== | |
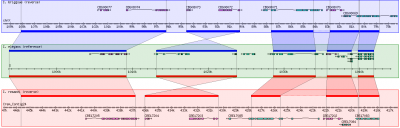
| − | + | [[Image:GBrowse_syn.png|none|400px|GBrowse_syn, as implemented at WormBase]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | == | + | ==Downloads== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | == | + | Download GBrowse_syn: [http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/] |
| − | + | ||
| − | + | ||
| − | + | The development version of GBrowse_syn is found at https://github.com/GMOD/GBrowse. Please be aware that development versions may have new features that are not fully tested. | |
| + | |||
| + | ==Using GBrowse_syn== | ||
| + | |||
| + | GBrowse_syn has been part of the [[GBrowse]] distribution since version 1.69; we recommend using the most up-to-date version of GBrowse 2. Please follow the [[GBrowse_2.0_Install_HOWTO|installation instructions for GBrowse]]. | ||
| + | |||
| + | ===Documentation=== | ||
| + | |||
| + | See the [[GBrowse_syn_Help|help for GBrowse_syn]] | ||
| + | |||
| + | ====Alignment data==== | ||
| − | |||
* GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments | * GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments | ||
| − | * There is an additional | + | * There is an additional GBrowse database for each species represented in the alignments |
| − | * The | + | * The databases for each species are configured in the same way as a regular GBrowse installations |
* [[GBrowse_syn_Database|Details on the GBrowse_syn database]] | * [[GBrowse_syn_Database|Details on the GBrowse_syn database]] | ||
| − | ==User interface== | + | ====User interface==== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | The overall look of Gbrowse_syn resembles GBrowse but has some key differences to accomodate the more complex comparative genome data (see the [[Screenshot|screenshot]] above). | |
| − | + | GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species. There is no upper limit to the number of species that can be displayed. | |
| − | == | + | ==Publications, Tutorials, and Presentations== |
| − | + | ===Publications on or mentioning GBrowse_syn=== | |
| + | |||
| + | Please refer to the following paper when citing GBrowse_syn: | ||
| + | |||
| + | * Using the Generic Synteny Browser (GBrowse_syn) <ref name=PMID:20836076/> | ||
| + | |||
| + | ===Tutorials=== | ||
| + | |||
| + | *[[GBrowse_syn_Tutorial|Installing and configuring GBrowse_syn]] | ||
| + | |||
| + | ===Presentations=== | ||
| − | |||
* [[:Media:GBrowse_syn_EBI2009.pdf|Challenges in Comparative Genome Browsing]] - Presented by [[User:Mckays|Sheldon McKay]] at the [http://www.ebi.ac.uk European Bioinformatics Institute], Hinxton, UK. | * [[:Media:GBrowse_syn_EBI2009.pdf|Challenges in Comparative Genome Browsing]] - Presented by [[User:Mckays|Sheldon McKay]] at the [http://www.ebi.ac.uk European Bioinformatics Institute], Hinxton, UK. | ||
* [[:Media:GBrowse_synSMBE2009.pdf|Comparative Genomics with GBrowse_syn]] - Presentation by [[User:Mckays|Sheldon McKay]] at the [http://ccg.biology.uiowa.edu/smbe/symposia.php?action=view&sym_ID=27 SMBE 2009 GMOD Workshop] on using [[GBrowse_syn]] for [[:Category:Comparative Genomics|comparative genomics]]. | * [[:Media:GBrowse_synSMBE2009.pdf|Comparative Genomics with GBrowse_syn]] - Presentation by [[User:Mckays|Sheldon McKay]] at the [http://ccg.biology.uiowa.edu/smbe/symposia.php?action=view&sym_ID=27 SMBE 2009 GMOD Workshop] on using [[GBrowse_syn]] for [[:Category:Comparative Genomics|comparative genomics]]. | ||
| Line 64: | Line 71: | ||
* [[Media:Gbrowse_syn.pdf|November 2007]] - [[User:Mckays|Sheldon McKay]]'s presentation on GBrowse_syn at the [[November 2007 GMOD Meeting#GBrowse_Syn|November 2007 GMOD Meeting]]. | * [[Media:Gbrowse_syn.pdf|November 2007]] - [[User:Mckays|Sheldon McKay]]'s presentation on GBrowse_syn at the [[November 2007 GMOD Meeting#GBrowse_Syn|November 2007 GMOD Meeting]]. | ||
| − | == | + | ==Contacts and Mailing Lists== |
| + | Support is via the GBrowse mailing list: | ||
| + | |||
| + | {| class="wikitable" | ||
| + | | | ||
| + | ! Mailing List Link | ||
| + | ! Description | ||
| + | ! Archive(s) | ||
| + | |- | ||
| + | ! rowspan="2" | [[GBrowse]] & [[GBrowse_syn]] | ||
| + | | [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse gmod-gbrowse] | ||
| + | | [[GBrowse]] and [[GBrowse_syn]] users and developers. | ||
| + | | [http://dir.gmane.org/gmane.science.biology.gmod.gbrowse Gmane], [http://gmod.827538.n3.nabble.com/GBrowse-f815907.html Nabble (2010/05+)], [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse Sourceforge] | ||
| + | |- | ||
| + | | [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse-cmts gmod-gbrowse-cmts] | ||
| + | | Code updates. | ||
| + | | [http://sourceforge.net/mailarchive/forum.php?forum_name=gmod-gbrowse-cmts Sourceforge] | ||
| + | |} | ||
| + | |||
| + | |||
| + | ==GBrowse_syn in the wild== | ||
| + | |||
| + | Public installations of GBrowse_syn: | ||
| + | *[http://www.arabidopsis.org/cgi-bin/gbrowse_syn/arabidopsis/?name=Chr1%3A8367000..8370501 The Arabidopsis Information Resource] | ||
| + | *[http://dev.wormbase.org/db/seq/gbrowse_syn/compara?search_src=Cele;name=X:1050001..1150000 WormBase] | ||
| + | *[http://solgenomics.net/gbrowse2/bin/gbrowse_syn/sol3/ Sol Genomics] | ||
| + | |||
| + | ==GBrowse_syn Development== | ||
| + | |||
| + | |||
| + | See the [[Talk:GBrowse_syn|discussion page]] for notes on further GBrowse_syn development. | ||
| + | |||
| + | |||
| + | ==See also== | ||
| + | |||
| + | The focus of this documentation is the GBrowse_syn application. However, the generation of whole genome alignments and identification of orthologous regions are the subject of considerable interest, so some background reading is listed below: | ||
| + | *[http://www.eecs.berkeley.edu/Pubs/TechRpts/2006/EECS-2006-104.html Primer on Hierarchical Genome Alignment Strategies] | ||
| + | *[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2577869/ article on PECAN and ENREDO] | ||
| + | *[http://www.ebi.ac.uk/~bjp/pecan/ all about PECAN] | ||
| + | *[http://www.ensembl.org/info/website/archives/index.html Information about EnsEMBL's compara pipeline] | ||
| + | |||
| + | ==More on GBrowse_syn== | ||
| + | |||
| + | See [[:Category:GBrowse_syn]] | ||
The [[:Image:GBrowse_syn_logo.png|GBrowse_syn logo]] was created by [mailto:NextLevelDesignStudios@gmail.com Darek Lakey], a participant in the [[Spring 2010 Logo Program]], while a design student at [http://www.linnbenton.edu Linn-Benton Community College]. | The [[:Image:GBrowse_syn_logo.png|GBrowse_syn logo]] was created by [mailto:NextLevelDesignStudios@gmail.com Darek Lakey], a participant in the [[Spring 2010 Logo Program]], while a design student at [http://www.linnbenton.edu Linn-Benton Community College]. | ||
| + | |||
| + | |||
| + | <references /> | ||
[[Category:GBrowse syn]] | [[Category:GBrowse syn]] | ||
Revision as of 16:03, 24 August 2012
__NOTITLE__
Contents
About Generic Synteny Browser (GBrowse_syn)
GBrowse_syn, or the Generic Synteny Browser, is a GBrowse-based synteny browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later).
Screenshots
Downloads
Download GBrowse_syn: http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/
The development version of GBrowse_syn is found at https://github.com/GMOD/GBrowse. Please be aware that development versions may have new features that are not fully tested.
Using GBrowse_syn
GBrowse_syn has been part of the GBrowse distribution since version 1.69; we recommend using the most up-to-date version of GBrowse 2. Please follow the installation instructions for GBrowse.
Documentation
See the help for GBrowse_syn
Alignment data
- GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments
- There is an additional GBrowse database for each species represented in the alignments
- The databases for each species are configured in the same way as a regular GBrowse installations
- Details on the GBrowse_syn database
User interface
The overall look of Gbrowse_syn resembles GBrowse but has some key differences to accomodate the more complex comparative genome data (see the screenshot above).
GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species. There is no upper limit to the number of species that can be displayed.
Publications, Tutorials, and Presentations
Publications on or mentioning GBrowse_syn
Please refer to the following paper when citing GBrowse_syn:
- Using the Generic Synteny Browser (GBrowse_syn) [1]
Tutorials
Presentations
- Challenges in Comparative Genome Browsing - Presented by Sheldon McKay at the European Bioinformatics Institute, Hinxton, UK.
- Comparative Genomics with GBrowse_syn - Presentation by Sheldon McKay at the SMBE 2009 GMOD Workshop on using GBrowse_syn for comparative genomics.
- GBrowse_syn at PAG - Presentation by Sheldon McKay at the Plant and Animal Genomes meeting, San Diego, CA, USA.
- November 2007 - Sheldon McKay's presentation on GBrowse_syn at the November 2007 GMOD Meeting.
Contacts and Mailing Lists
Support is via the GBrowse mailing list:
| Mailing List Link | Description | Archive(s) | |
|---|---|---|---|
| GBrowse & GBrowse_syn | gmod-gbrowse | GBrowse and GBrowse_syn users and developers. | Gmane, Nabble (2010/05+), Sourceforge |
| gmod-gbrowse-cmts | Code updates. | Sourceforge |
GBrowse_syn in the wild
Public installations of GBrowse_syn:
GBrowse_syn Development
See the discussion page for notes on further GBrowse_syn development.
See also
The focus of this documentation is the GBrowse_syn application. However, the generation of whole genome alignments and identification of orthologous regions are the subject of considerable interest, so some background reading is listed below:
- Primer on Hierarchical Genome Alignment Strategies
- article on PECAN and ENREDO
- all about PECAN
- Information about EnsEMBL's compara pipeline
More on GBrowse_syn
See Category:GBrowse_syn The GBrowse_syn logo was created by Darek Lakey, a participant in the Spring 2010 Logo Program, while a design student at Linn-Benton Community College.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedPMID:20836076