| Available on platform | web + |
| Date published | 2009 + |
| Has DOI | 10.1093/bioinformatics/btp458 + |
| Has PMCID | PMC2773250 + |
| Has PMID | 19648141 + |
| Has URL | http://sourceforge.net/projects/gmod/files/cmap/ +, https://gmod.svn.sourceforge.net/svnroot/gmod/cmap +, http://www.gramene.org/db/cmap +, http://www.gramene.org/cmap +, http://www.comparative-legumes.org/ +, http://racerx00.tamu.edu/cgi-bin/cmap/viewer +, http://bioinformatics.pbcbasc.latrobe.edu.au/basc/cgi-bin/index.cgi +, http://rye.pw.usda.gov/cgi-bin/cmap/map_set_info?map_set_aid=Barley_consensus_2003 +, http://magi.plantgenomics.iastate.edu/cgi-bin/cmap/map_set_info +, http://www.bioinfo.wsu.edu/cgi-bin/gdr/cmap/map_set_info?map_set_aid=17 +, http://www.cottonmarker.org/ +, http://bioinf.scri.sari.ac.uk/ +, http://www.pristionchus.org/cgi-bin/cmap/viewer?ref_map_set_aid=PM%3Bref_map_aids=109 +, http://medvet.angis.org.au/cmap/ +, http://soybeanbreederstoolbox.org/index.php +, http://map.lab.nig.ac.jp:8080/cmap/ +, http://peanutgenetics.tamu.edu/cmap/ +, http://www.cottondb.org/cmap/ +, http://tropgenedb.cirad.fr + and http://cricket.ornl.gov/cgi-bin/cmap/map_set_info + |
| Has author | Youens-Clark K +, Faga B +, Yap IV +, Stein L + and Ware D + |
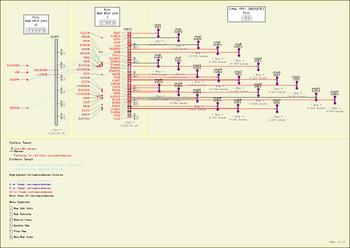
| Has description | Comparative mapping resource for grasses (plant) +, A publicly accessible legume resource that will integrate genetic and molecular data from multiple legume species and enable cross-species comparisons +, The Honey Bee genome database +, Brassica database + and A browser-based tool for the visual comparison of various maps (sequence, genetic, etc.) from any number of species. The CMap package also includes tools for curating map data. + |
| Has development status | inactive + |
| Has input format | tab-delimited + and GFF + |
| Has licence | GPL + |
| Has logo | CMapLogo-horiz.png + |
| Has output format | tab-delimited + |
| Has publication details | Bioinformatics (Oxford, England) 25: 3040-2 + |
| Has software maturity status | mature + |
| Has support status | active + |
| Has title | Gramene +, NCGR's Legume Information System +, BeeBase +, BASC +, GrainGenes +, MAGI +, Genome Database for Rosaceae +, Cotton Marker Database +, SCRI Plant Bioinformatics Group +, Pristionchus pacificus Physical Map +, Comparitive Location Database +, Soybean Breeders Toolbox +, Composite Wheat Map +, PeanutMap +, CottonDB +, TropGene +, Populus at Oak Ridge + and CMap 1.01: a comparative mapping application for the Internet. + |
| Has topic | CMap + |
| Is open source | Yes + |
| Link type | download +, source code +, demo server +, public server + and wild URL + |
| Published in | Bioinformatics (Oxford, England) + |
| Release date | 2002 + |
| Tool functionality or classification | Genome Visualization & Editing +, Comparative Genome Visualization + and Database schema + |
| Written in language | Perl + |
| Has subobjectThis property is a special property in this wiki. | CMap#http://sourceforge.net/projects/gmod/files/cmap/ +, CMap#https://gmod.svn.sourceforge.net/svnroot/gmod/cmap +, CMap#http://www.gramene.org/db/cmap +, CMap#http://www.gramene.org/cmap +, CMap#http://www.comparative-legumes.org/ +, CMap#http://racerx00.tamu.edu/cgi-bin/cmap/viewer +, CMap#http://bioinformatics.pbcbasc.latrobe.edu.au/basc/cgi-bin/index.cgi +, CMap#http://rye.pw.usda.gov/cgi-bin/cmap/map_set_info?map_set_aid=Barley_consensus_2003 +, CMap#http://magi.plantgenomics.iastate.edu/cgi-bin/cmap/map_set_info +, CMap#http://www.bioinfo.wsu.edu/cgi-bin/gdr/cmap/map_set_info?map_set_aid=17 +, CMap#http://www.cottonmarker.org/ +, CMap#http://bioinf.scri.sari.ac.uk/ +, CMap#http://www.pristionchus.org/cgi-bin/cmap/viewer?ref_map_set_aid=PM;ref_map_aids=109 +, CMap#http://medvet.angis.org.au/cmap/ +, CMap#http://soybeanbreederstoolbox.org/index.php +, CMap#http://map.lab.nig.ac.jp:8080/cmap/ +, CMap#http://peanutgenetics.tamu.edu/cmap/ +, CMap#http://www.cottondb.org/cmap/ +, CMap#http://tropgenedb.cirad.fr + and CMap#http://cricket.ornl.gov/cgi-bin/cmap/map_set_info + |